6O9M
 
 | | Structure of the human apo TFIIH | | Descriptor: | CDK-activating kinase assembly factor MAT1, General transcription factor IIH subunit 1, General transcription factor IIH subunit 2, ... | | Authors: | Yan, C.L, Dodd, T, He, Y, Tainer, J.A, Tsutakawa, S.E, Ivanov, I. | | Deposit date: | 2019-03-14 | | Release date: | 2019-05-29 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (4.4 Å) | | Cite: | Transcription preinitiation complex structure and dynamics provide insight into genetic diseases.
Nat.Struct.Mol.Biol., 26, 2019
|
|
8EZB
 
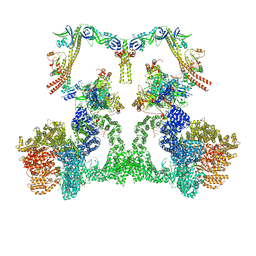 | | NHEJ Long-range complex with ATP | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, DNA (30-MER), DNA (31-MER), ... | | Authors: | Chen, S, He, Y. | | Deposit date: | 2022-10-31 | | Release date: | 2023-06-14 | | Method: | ELECTRON MICROSCOPY (8.9 Å) | | Cite: | Cryo-EM visualization of DNA-PKcs structural intermediates in NHEJ.
Sci Adv, 9, 2023
|
|
8EZA
 
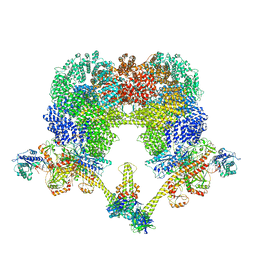 | | NHEJ Long-range complex with PAXX | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, DNA (30-MER), DNA (31-MER), ... | | Authors: | Chen, S, He, Y. | | Deposit date: | 2022-10-31 | | Release date: | 2023-06-14 | | Method: | ELECTRON MICROSCOPY (4.39 Å) | | Cite: | Cryo-EM visualization of DNA-PKcs structural intermediates in NHEJ.
Sci Adv, 9, 2023
|
|
8EZ9
 
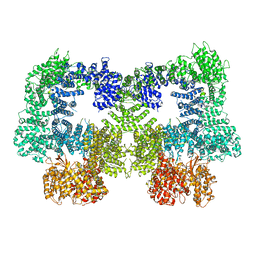 | | Dimeric complex of DNA-PKcs | | Descriptor: | DNA-dependent protein kinase catalytic subunit, unknown region of DNA-PKcs | | Authors: | Chen, S, He, Y. | | Deposit date: | 2022-10-31 | | Release date: | 2023-06-14 | | Method: | ELECTRON MICROSCOPY (5.67 Å) | | Cite: | Cryo-EM visualization of DNA-PKcs structural intermediates in NHEJ.
Sci Adv, 9, 2023
|
|
5DGY
 
 | | Crystal structure of rhodopsin bound to visual arrestin | | Descriptor: | Endolysin,Rhodopsin,S-arrestin | | Authors: | Zhou, X.E, Gao, X, Kang, Y, He, Y, de Waal, P.W, Suino-Powell, K.M, Wang, M, Melcher, K, Xu, H.E. | | Deposit date: | 2015-08-28 | | Release date: | 2016-03-23 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (7.7 Å) | | Cite: | X-ray laser diffraction for structure determination of the rhodopsin-arrestin complex.
Sci Data, 3, 2016
|
|
8GA8
 
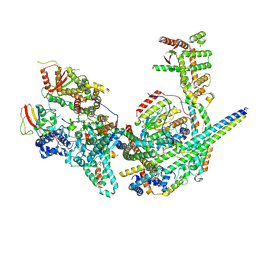 | | Structure of the yeast (HDAC) Rpd3L complex | | Descriptor: | Histone deacetylase RPD3, Transcriptional regulatory protein DEP1, Transcriptional regulatory protein PHO23, ... | | Authors: | Patel, A.B, Radhakrishnan, I, He, Y. | | Deposit date: | 2023-02-22 | | Release date: | 2023-07-12 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Cryo-EM structure of the Saccharomyces cerevisiae Rpd3L histone deacetylase complex.
Nat Commun, 14, 2023
|
|
4LYC
 
 | | Cd ions within a lysoyzme single crystal | | Descriptor: | CADMIUM ION, Lysozyme C | | Authors: | Wei, H, House, S, Wu, J, Zhang, J, Wang, Z, He, Y, Gao, Y.-G, Robinson, H, Li, W, Zuo, J.-M, Robertson, I.M, Lu, Y. | | Deposit date: | 2013-07-30 | | Release date: | 2015-02-25 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Enhanced and tunable fluorescent quantum dots within a single crystal of protein
TO BE PUBLISHED
|
|
4LYB
 
 | | CdS within a lysoyzme single crystal | | Descriptor: | CADMIUM ION, Lysozyme C | | Authors: | Wei, H, House, S, Wu, J, Zhang, J, Wang, Z, He, Y, Gao, Y.-G, Robinson, H, Li, W, Zuo, J.-M, Robertson, I.M, Lu, Y. | | Deposit date: | 2013-07-30 | | Release date: | 2015-02-25 | | Method: | X-RAY DIFFRACTION (1.21 Å) | | Cite: | Enhanced and tunable fluorescent quantum dots within a single crystal of protein
TO BE PUBLISHED
|
|
6CND
 
 | | Yeast RNA polymerase III natural open complex (nOC) | | Descriptor: | DNA (71-MER), DNA-directed RNA polymerase III subunit RPC1, DNA-directed RNA polymerase III subunit RPC10, ... | | Authors: | Han, Y, He, Y. | | Deposit date: | 2018-03-08 | | Release date: | 2018-08-22 | | Last modified: | 2020-01-08 | | Method: | ELECTRON MICROSCOPY (4.8 Å) | | Cite: | Structural visualization of RNA polymerase III transcription machineries.
Cell Discov, 4, 2018
|
|
6CNB
 
 | | Yeast RNA polymerase III initial transcribing complex | | Descriptor: | DNA (71-MER), DNA-directed RNA polymerase III subunit RPC1, DNA-directed RNA polymerase III subunit RPC10, ... | | Authors: | Han, Y, He, Y. | | Deposit date: | 2018-03-08 | | Release date: | 2018-08-22 | | Last modified: | 2020-01-08 | | Method: | ELECTRON MICROSCOPY (4.1 Å) | | Cite: | Structural visualization of RNA polymerase III transcription machineries.
Cell Discov, 4, 2018
|
|
6CNC
 
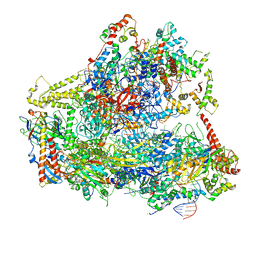 | | Yeast RNA polymerase III open complex | | Descriptor: | DNA (71-MER), DNA-directed RNA polymerase III subunit RPC1, DNA-directed RNA polymerase III subunit RPC10, ... | | Authors: | Han, Y, He, Y. | | Deposit date: | 2018-03-08 | | Release date: | 2018-08-22 | | Last modified: | 2020-01-08 | | Method: | ELECTRON MICROSCOPY (4.1 Å) | | Cite: | Structural visualization of RNA polymerase III transcription machineries.
Cell Discov, 4, 2018
|
|
5ZMM
 
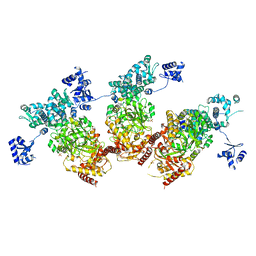 | | Structure of the Type IV phosphorothioation-dependent restriction endonuclease ScoMcrA | | Descriptor: | SULFATE ION, Uncharacterized protein McrA, ZINC ION | | Authors: | Liu, G, Fu, W, Zhang, Z, He, Y, Yu, H, Zhao, Y, Deng, Z, Wu, G, He, X. | | Deposit date: | 2018-04-04 | | Release date: | 2018-09-26 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (3.15 Å) | | Cite: | Structural basis for the recognition of sulfur in phosphorothioated DNA.
Nat Commun, 9, 2018
|
|
5ZMN
 
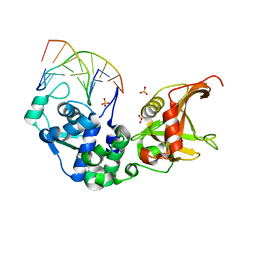 | | Sulfur binding domain and SRA domain of ScoMcrA complexed with phosphorothioated DNA | | Descriptor: | DNA (5'-D(*CP*CP*CP*GP*(GS)P*CP*CP*GP*GP*G)-3'), SULFATE ION, Uncharacterized protein McrA | | Authors: | Liu, G, Fu, W, Zhang, Z, He, Y, Yu, H, Zhao, Y, Deng, Z, Wu, G, He, X. | | Deposit date: | 2018-04-04 | | Release date: | 2018-09-26 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (3.29 Å) | | Cite: | Structural basis for the recognition of sulfur in phosphorothioated DNA.
Nat Commun, 9, 2018
|
|
3L8J
 
 | | Crystal structure of CCM3, a cerebral cavernous malformation protein critical for vascular integrity | | Descriptor: | Programmed cell death protein 10 | | Authors: | Li, X, Zhang, R, Zhang, H, He, Y, Ji, W, Min, W, Boggon, T.J. | | Deposit date: | 2009-12-31 | | Release date: | 2010-05-19 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (3.05 Å) | | Cite: | Crystal structure of CCM3, a cerebral cavernous malformation protein critical for vascular integrity.
J.Biol.Chem., 285, 2010
|
|
3L8I
 
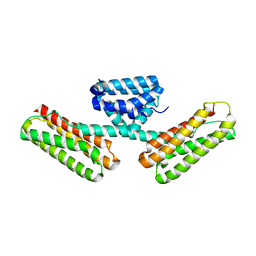 | | Crystal structure of CCM3, a cerebral cavernous malformation protein critical for vascular integrity | | Descriptor: | Programmed cell death protein 10 | | Authors: | Li, X, Zhang, R, Zhang, H, He, Y, Ji, W, Min, W, Boggon, T.J. | | Deposit date: | 2009-12-31 | | Release date: | 2010-05-19 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of CCM3, a cerebral cavernous malformation protein critical for vascular integrity.
J.Biol.Chem., 285, 2010
|
|
2KFW
 
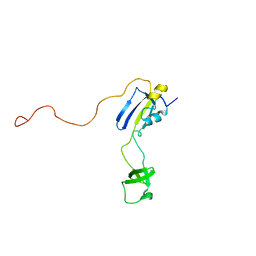 | | Solution structure of full-length SlyD from E.coli | | Descriptor: | FKBP-type peptidyl-prolyl cis-trans isomerase slyD | | Authors: | Martino, L, He, Y, Hands-Taylor, K.L, Valentine, E.R, Kelly, G, Giancola, C, Conte, M.R. | | Deposit date: | 2009-02-28 | | Release date: | 2009-09-15 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | The interaction of the Escherichia coli protein SlyD with nickel ions illuminates the mechanism of regulation of its peptidyl-prolyl isomerase activity.
Febs J., 276, 2009
|
|
6V85
 
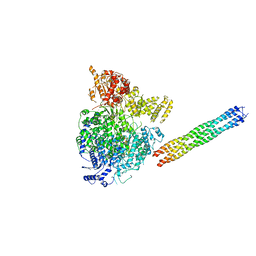 | | Parainfluenza virus 5 L-P complex | | Descriptor: | Phosphoprotein, RNA-directed RNA polymerase L, ZINC ION | | Authors: | Abdella, R, He, Y. | | Deposit date: | 2019-12-10 | | Release date: | 2020-02-19 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (4.38 Å) | | Cite: | Structure of a paramyxovirus polymerase complex reveals a unique methyltransferase-CTD conformation.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
6V86
 
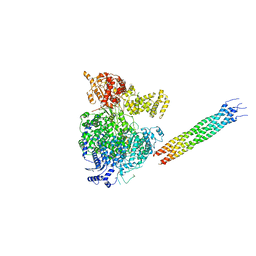 | |
6VAG
 
 | | Crystal structure of the oligomerization domain of phosphoprotein from parainfluenza virus 5 | | Descriptor: | GLYCEROL, Phosphoprotein | | Authors: | Aggarwal, M, Abdella, R, He, Y, Lamb, R.A. | | Deposit date: | 2019-12-17 | | Release date: | 2020-02-19 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Structure of a paramyxovirus polymerase complex reveals a unique methyltransferase-CTD conformation.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
2LD7
 
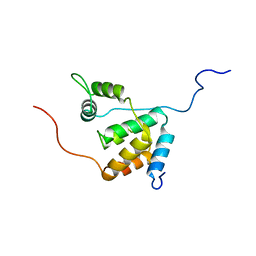 | | Solution structure of the mSin3A PAH3-SAP30 SID complex | | Descriptor: | Histone deacetylase complex subunit SAP30, Paired amphipathic helix protein Sin3a | | Authors: | Xie, T, He, Y, Korkeamaki, H, Zhang, Y, Imhoff, R, Lohi, O, Radhakrishnan, I. | | Deposit date: | 2011-05-16 | | Release date: | 2011-06-15 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Structure of the 30-kDa Sin3-associated protein (SAP30) in complex with the mammalian Sin3A corepressor and its role in nucleic acid binding.
J.Biol.Chem., 286, 2011
|
|
7LBM
 
 | | Structure of the human Mediator-bound transcription pre-initiation complex | | Descriptor: | CDK-activating kinase assembly factor MAT1, Cyclin-H, Cyclin-dependent kinase 7, ... | | Authors: | Abdella, R, Talyzina, A, He, Y. | | Deposit date: | 2021-01-08 | | Release date: | 2021-03-24 | | Last modified: | 2021-04-14 | | Method: | ELECTRON MICROSCOPY (4.8 Å) | | Cite: | Structure of the human Mediator-bound transcription preinitiation complex.
Science, 372, 2021
|
|
2KQ1
 
 | | Solution Structure Of Protein BH0266 From Bacillus halodurans. Northeast Structural Genomics Consortium Target BhR97a | | Descriptor: | BH0266 protein | | Authors: | Wu, Y, Eletsky, A, Lee, D, Sathyamoorthy, B, Buchwald, W, Hua, J, Ciccosanti, C, He, Y, Hamilton, K, Acton, T, Xiao, R, Everett, J, Lee, H, Prestegard, J, Montelione, G.T, Szyperski, T, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2009-10-23 | | Release date: | 2009-12-22 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Solution Structure Of Protein BH0266 From Bacillus halodurans
To be Published
|
|
5Y14
 
 | | Crystal structure of LP-40/N44 | | Descriptor: | LP-40, N44 | | Authors: | Zhang, X, Wang, X, He, Y. | | Deposit date: | 2017-07-19 | | Release date: | 2017-11-08 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.762 Å) | | Cite: | Enfuvirtide (T20)-Based Lipopeptide Is a Potent HIV-1 Cell Fusion Inhibitor: Implications for Viral Entry and Inhibition
J. Virol., 91, 2017
|
|
5YB3
 
 | | Crystal structure of HP23L/N36 | | Descriptor: | Envelope glycoprotein, HP23L | | Authors: | Zhang, X, Wang, X, He, Y. | | Deposit date: | 2017-09-03 | | Release date: | 2018-02-28 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.043 Å) | | Cite: | Structural Insights into the Mechanisms of Action of Short-Peptide HIV-1 Fusion Inhibitors Targeting the Gp41 Pocket
Front Cell Infect Microbiol, 8, 2018
|
|
5YB2
 
 | | Crystal structure of LP-11/N44 | | Descriptor: | Envelope glycoprotein, LP-11 | | Authors: | Zhang, X, Wang, X, He, Y. | | Deposit date: | 2017-09-03 | | Release date: | 2018-02-28 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (3.8 Å) | | Cite: | Structural Insights into the Mechanisms of Action of Short-Peptide HIV-1 Fusion Inhibitors Targeting the Gp41 Pocket
Front Cell Infect Microbiol, 8, 2018
|
|
