2VUK
 
 | |
8BZP
 
 | | JNK3 (Mitogen-activated protein kinase 10) in Complex with Compound 23 bearing a C(sp3)F2Br moiety | | Descriptor: | 1,2-ETHANEDIOL, 2-bromanyl-2,2-bis(fluoranyl)-~{N}-(5-pyridin-4-yl-1,3,4-thiadiazol-2-yl)ethanamide, BETA-MERCAPTOETHANOL, ... | | Authors: | Stahlecker, J, Vaas, S, Stehle, T, Boeckler, F.M. | | Deposit date: | 2022-12-15 | | Release date: | 2023-08-02 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | Principles and Applications of CF 2 X Moieties as Unconventional Halogen Bond Donors in Medicinal Chemistry, Chemical Biology, and Drug Discovery.
J.Med.Chem., 66, 2023
|
|
7ZH8
 
 | | DYRK1a in Complex with a Bromo-Triazolo-Pyridine | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, 6-bromanyl-3H-[1,2,3]triazolo[4,5-b]pyridine, CHLORIDE ION, ... | | Authors: | Dammann, M, Stahlecker, J, Stehle, T, Boeckler, F.M. | | Deposit date: | 2022-04-05 | | Release date: | 2022-11-09 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Screening of a Halogen-Enriched Fragment Library Leads to Unconventional Binding Modes.
J.Med.Chem., 65, 2022
|
|
8A92
 
 | | p53-Y220C Core Domain in Complex with a Bromo-trifluoro-pyrazole-amine | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, 4-bromanyl-5-(trifluoromethyl)-1H-pyrazol-3-amine, Cellular tumor antigen p53, ... | | Authors: | Stahlecker, J, Braun, M.B, Stehle, T, Boeckler, F.M. | | Deposit date: | 2022-06-27 | | Release date: | 2022-11-09 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.37 Å) | | Cite: | Revisiting a challenging p53 binding site: a diversity-optimized HEFLib reveals diverse binding modes in T-p53C-Y220C.
Rsc Med Chem, 13, 2022
|
|
5AOM
 
 | | Structure of the p53 cancer mutant Y220C with bound small molecule PhiKan883 | | Descriptor: | CELLULAR TUMOR ANTIGEN P53, GLYCEROL, N-(5-chloranyl-2-oxidanyl-phenyl)piperidine-4-carboxamide, ... | | Authors: | Joerger, A.C, Boeckler, F.M, Wilcken, R. | | Deposit date: | 2015-09-10 | | Release date: | 2015-12-16 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.74 Å) | | Cite: | Exploiting Transient Protein States for the Design of Small-Molecule Stabilizers of Mutant P53.
Structure, 23, 2015
|
|
4AGM
 
 | | Structure of the p53 core domain mutant Y220C bound to the stabilizing small molecule PhiKan5086 | | Descriptor: | 2-{[4-(DIETHYLAMINO)PIPERIDIN-1-YL]METHYL}-4,6-DIIODOPHENOL, CELLULAR TUMOR ANTIGEN P53, ZINC ION | | Authors: | Joerger, A.C, Wilcken, R, Boeckler, F.M, Fersht, A.R. | | Deposit date: | 2012-01-30 | | Release date: | 2012-03-21 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.52 Å) | | Cite: | Halogen-Enriched Fragment Libraries as Leads for Drug Rescue of Mutant P53.
J.Am.Chem.Soc., 134, 2012
|
|
4AGN
 
 | | Structure of the p53 core domain mutant Y220C bound to the stabilizing small molecule PhiKan5116 | | Descriptor: | 2-{[4-(DIETHYLAMINO)PIPERIDIN-1-YL]METHYL}-4-(3-HYDROXYPROP-1-YN-1-YL)-6-IODOPHENOL, CELLULAR TUMOR ANTIGEN P53, ZINC ION | | Authors: | Joerger, A.C, Wilcken, R, Boeckler, F.M, Fersht, A.R. | | Deposit date: | 2012-01-30 | | Release date: | 2012-03-21 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Halogen-Enriched Fragment Libraries as Leads for Drug Rescue of Mutant P53.
J.Am.Chem.Soc., 134, 2012
|
|
4AGP
 
 | | Structure of the p53 core domain mutant Y220C bound to the stabilizing small molecule PhiKan5176 | | Descriptor: | 2-{[4-(diethylamino)piperidin-1-yl]methyl}-6-iodo-4-(3-phenoxyprop-1-yn-1-yl)phenol, CELLULAR TUMOR ANTIGEN P53, ZINC ION | | Authors: | Joerger, A.C, Wilcken, R, Fersht, A.R, Boeckler, F.M. | | Deposit date: | 2012-01-30 | | Release date: | 2012-03-21 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Halogen-Enriched Fragment Libraries as Leads for Drug Rescue of Mutant P53.
J.Am.Chem.Soc., 134, 2012
|
|
4AGO
 
 | | Structure of the p53 core domain mutant Y220C bound to the stabilizing small molecule PhiKan5174 | | Descriptor: | CELLULAR TUMOR ANTIGEN P53, TERT-BUTYL [3-(3-{[4-(DIETHYLAMINO)PIPERIDIN-1-YL]METHYL}-4-HYDROXY-5-IODOPHENYL)PROP-2-YN-1-YL]CARBAMATE, ZINC ION | | Authors: | Joerger, A.C, Wilcken, R, Fersht, A.R, Boeckler, F.M. | | Deposit date: | 2012-01-30 | | Release date: | 2012-03-21 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Halogen-Enriched Fragment Libraries as Leads for Drug Rescue of Mutant P53.
J.Am.Chem.Soc., 134, 2012
|
|
4AGL
 
 | | Structure of the p53 core domain mutant Y220C bound to the stabilizing small molecule PhiKan784 | | Descriptor: | 2,4-BIS(IODANYL)-6-[[METHYL-(1-METHYLPIPERIDIN-4-YL)AMINO]METHYL]PHENOL, CELLULAR TUMOR ANTIGEN P53, ZINC ION | | Authors: | Joerger, A.C, Wilcken, R, Boeckler, F.M, Fersht, A.R. | | Deposit date: | 2012-01-30 | | Release date: | 2012-03-21 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Halogen-Enriched Fragment Libraries as Leads for Drug Rescue of Mutant P53.
J.Am.Chem.Soc., 134, 2012
|
|
4AGQ
 
 | | Structure of the p53 core domain mutant Y220C bound to the stabilizing small molecule PhiKan5196 | | Descriptor: | 2-{[4-(diethylamino)piperidin-1-yl]methyl}-6-iodo-4-[3-(phenylamino)prop-1-yn-1-yl]phenol, CELLULAR TUMOR ANTIGEN P53, ZINC ION | | Authors: | Joerger, A.C, Wilcken, R, Boeckler, F.M, Fersht, A.R. | | Deposit date: | 2012-01-30 | | Release date: | 2012-03-21 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.42 Å) | | Cite: | Halogen-Enriched Fragment Libraries as Leads for Drug Rescue of Mutant P53.
J.Am.Chem.Soc., 134, 2012
|
|
4X21
 
 | | The MAP kinase JNK3 as target for halogen bonding | | Descriptor: | CHLORIDE ION, Mitogen-activated protein kinase 10, N-ethyl-4-{[4-(1H-indol-3-yl)-5-iodopyrimidin-2-yl]amino}piperidine-1-carboxamide | | Authors: | Lange, A, Buettner, F.M, Guenther, M.B, Zimmermann, M.O, Hennig, S, Laufer, S.A, Stehle, T, Boeckler, F. | | Deposit date: | 2014-11-25 | | Release date: | 2015-11-11 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Targeting the Gatekeeper MET146 of C-Jun N-Terminal Kinase 3 Induces a Bivalent Halogen/Chalcogen Bond.
J.Am.Chem.Soc., 137, 2015
|
|
6QJU
 
 | |
3UVQ
 
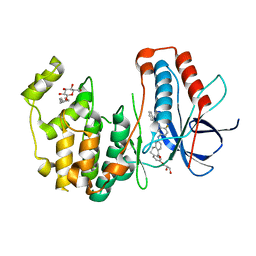 | | Human p38 MAP Kinase in Complex with a Dibenzosuberone Derivative | | Descriptor: | Mitogen-activated protein kinase 14, N-{5-[(7-{[(2R)-2,3-dihydroxypropyl]oxy}-5-oxo-10,11-dihydro-5H-dibenzo[a,d][7]annulen-2-yl)amino]-2-fluorophenyl}benzamide, octyl beta-D-glucopyranoside | | Authors: | Mayer-Wrangowski, S.C, Richters, A, Gruetter, C, Rauh, D. | | Deposit date: | 2011-11-30 | | Release date: | 2012-12-05 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Dibenzosuberones as p38 mitogen-activated protein kinase inhibitors with low ATP competitiveness and outstanding whole blood activity.
J.Med.Chem., 56, 2013
|
|
5A7B
 
 | |
5AB9
 
 | |
5ABA
 
 | |
5G4O
 
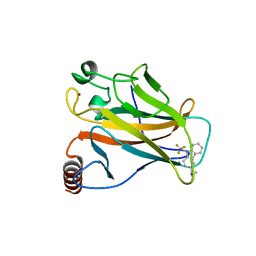 | | Crystal structure of the p53 cancer mutant Y220C in complex with a trifluorinated derivative of the small molecule stabilizer Phikan083 | | Descriptor: | CELLULAR TUMOR ANTIGEN P53, N,N-dimethyl-1-[9-(2,2,2-trifluoroethyl)-9H-carbazol-3-yl]methanamine, ZINC ION | | Authors: | Joerger, A.C, Bauer, M, Baud, M.G.J, Spencer, J. | | Deposit date: | 2016-05-13 | | Release date: | 2016-06-22 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.48 Å) | | Cite: | Harnessing Fluorine-Sulfur Contacts and Multipolar Interactions for the Design of P53 Mutant Y220C Rescue Drugs.
Acs Chem.Biol., 11, 2016
|
|
5G4N
 
 | | Crystal structure of the p53 cancer mutant Y220C in complex with a difluorinated derivative of the small molecule stabilizer Phikan083 | | Descriptor: | 1-[9-(2,2-difluoroethyl)-9H-carbazol-3-yl]-N-methylmethanamine, CELLULAR TUMOR ANTIGEN P53, GLYCEROL, ... | | Authors: | Joerger, A.C, Bauer, M, Jones, R.N, Spencer, J. | | Deposit date: | 2016-05-13 | | Release date: | 2016-06-22 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Harnessing Fluorine-Sulfur Contacts and Multipolar Interactions for the Design of P53 Mutant Y220C Rescue Drugs.
Acs Chem.Biol., 11, 2016
|
|
5G4M
 
 | |
6EQ9
 
 | | Crystal structure of JNK3 in complex with AMP-PCP | | Descriptor: | BETA-MERCAPTOETHANOL, CHLORIDE ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Macedo, J.T, Stehle, T, Blaum, B.S. | | Deposit date: | 2017-10-12 | | Release date: | 2018-08-08 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.83 Å) | | Cite: | Structural Optimization of a Pyridinylimidazole Scaffold: Shifting the Selectivity from p38 alpha Mitogen-Activated Protein Kinase to c-Jun N-Terminal Kinase 3.
ACS Omega, 3, 2018
|
|
6EKD
 
 | | Crystal structure of JNK3 in complex with a pyridinylimidazole inhibitor | | Descriptor: | 1,2-ETHANEDIOL, 4-(4-methyl-2-methylsulfanyl-1~{H}-imidazol-5-yl)-~{N}-(4-morpholin-4-ylphenyl)pyridin-2-amine, BETA-MERCAPTOETHANOL, ... | | Authors: | Macedo, J.T, Stehle, T, Blaum, B.S. | | Deposit date: | 2017-09-26 | | Release date: | 2018-08-08 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural Optimization of a Pyridinylimidazole Scaffold: Shifting the Selectivity from p38 alpha Mitogen-Activated Protein Kinase to c-Jun N-Terminal Kinase 3.
ACS Omega, 3, 2018
|
|
6EMH
 
 | | Crystal structure of JNK3 in complex with a pyridinylimidazole inhibitor | | Descriptor: | 1,2-ETHANEDIOL, 4-(4-methyl-1~{H}-imidazol-5-yl)-~{N}-(4-morpholin-4-ylphenyl)pyridin-2-amine, BETA-MERCAPTOETHANOL, ... | | Authors: | Macedo, J.T, Stehle, T, Blaum, B.S. | | Deposit date: | 2017-10-02 | | Release date: | 2018-08-08 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | Structural Optimization of a Pyridinylimidazole Scaffold: Shifting the Selectivity from p38 alpha Mitogen-Activated Protein Kinase to c-Jun N-Terminal Kinase 3.
ACS Omega, 3, 2018
|
|
5AOL
 
 | |
5AOJ
 
 | | Structure of the p53 cancer mutant Y220C in complex with 2-hydroxy-3, 5-diiodo-4-(1H-pyrrol-1-yl)benzoic acid | | Descriptor: | 2-hydroxy-3,5-diiodo-4-(1H-pyrrol-1-yl)benzoic acid, CELLULAR TUMOR ANTIGEN P53, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Joerger, A.C, Baud, M.G, Bauer, M.R, Fersht, A.R. | | Deposit date: | 2015-09-10 | | Release date: | 2015-12-16 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.47 Å) | | Cite: | Exploiting Transient Protein States for the Design of Small-Molecule Stabilizers of Mutant P53.
Structure, 23, 2015
|
|
