3KD2
 
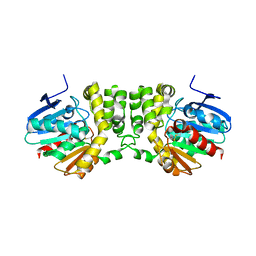 | | Crystal structure of the CFTR inhibitory factor Cif | | Descriptor: | CFTR inhibitory factor (Cif) | | Authors: | Bahl, C.D, Madden, D.R. | | Deposit date: | 2009-10-22 | | Release date: | 2010-01-26 | | Last modified: | 2011-10-12 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of the cystic fibrosis transmembrane conductance regulator inhibitory factor Cif reveals novel active-site features of an epoxide hydrolase virulence factor.
J.Bacteriol., 192, 2010
|
|
4YX9
 
 | |
3PI6
 
 | |
3KDA
 
 | |
4MEA
 
 | |
4DMF
 
 | |
4DNF
 
 | |
4DMC
 
 | |
4DNO
 
 | | Crystal structure of the CFTR inhibitory factor Cif with the E153Q mutation adducted with the 1,2-epoxyhexane hydrolysis intermediate | | Descriptor: | (2R)-hexane-1,2-diol, Putative hydrolase | | Authors: | Bahl, C.D, Patankar, Y.R, Madden, D.R. | | Deposit date: | 2012-02-08 | | Release date: | 2013-08-07 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Visualizing the Mechanism of Epoxide Hydrolysis by the Bacterial Virulence Enzyme Cif.
Biochemistry, 55, 2016
|
|
4DLN
 
 | |
4DM7
 
 | |
4EUS
 
 | |
4DMK
 
 | |
4DMH
 
 | |
4MEB
 
 | | Crystal structure of aCif-D158S | | Descriptor: | GLYCEROL, PHOSPHATE ION, Predicted protein | | Authors: | Bridges, A.A, Bahl, C.D, Madden, D.R. | | Deposit date: | 2013-08-25 | | Release date: | 2014-02-05 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Signature motifs identify an acinetobacter cif virulence factor with epoxide hydrolase activity.
J.Biol.Chem., 289, 2014
|
|
4EHB
 
 | |
5W9F
 
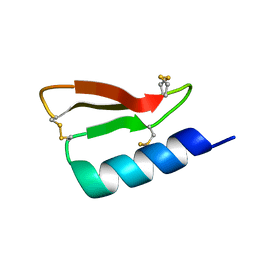 | | Solution structure of the de novo mini protein gHEEE_02 | | Descriptor: | De novo mini protein gHEEE_02 | | Authors: | Pulavarti, S.V.S.R.K, Shaw, E.A, Bahl, C.D, Garry, B.W, Baker, D, Szyperski, T. | | Deposit date: | 2017-06-23 | | Release date: | 2018-07-11 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Cytosolic expression, solution structures, and molecular dynamics simulation of genetically encodable disulfide-rich de novo designed peptides.
Protein Sci., 27, 2018
|
|
2ND3
 
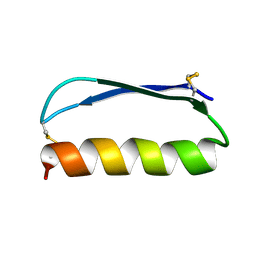 | | Solution structure of the de novo mini protein gEEH_04 | | Descriptor: | De novo mini protein EEH_04 | | Authors: | Pulavarti, S.V, Bahl, C.D, Gilmore, J.M, Eletsky, A, Buchko, G.W, Baker, D, Szyperski, T. | | Deposit date: | 2016-04-22 | | Release date: | 2016-09-21 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Accurate de novo design of hyperstable constrained peptides.
Nature, 538, 2016
|
|
2ND2
 
 | | Solution structure of the de novo mini protein gHHH_06 | | Descriptor: | De novo mini protein HHH_06 | | Authors: | Pulavarti, S.V, Eletsky, A, Bahl, C.D, Buchko, G.W, Baker, D, Szyperski, T. | | Deposit date: | 2016-04-22 | | Release date: | 2016-09-21 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Accurate de novo design of hyperstable constrained peptides.
Nature, 538, 2016
|
|
5JI4
 
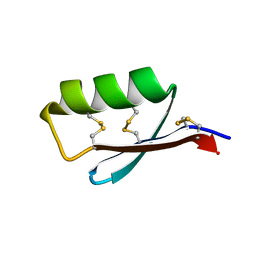 | | Solution structure of the de novo mini protein gEEHE_02 | | Descriptor: | W37 | | Authors: | Buchko, G.W, Bahl, C.D, Pulavarti, S.V, Baker, D, Szyperski, T. | | Deposit date: | 2016-04-21 | | Release date: | 2016-09-28 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Accurate de novo design of hyperstable constrained peptides.
Nature, 538, 2016
|
|
5JHI
 
 | | Solution structure of the de novo mini protein gEHE_06 | | Descriptor: | W35 | | Authors: | Buchko, G.W, Bahl, C.D, Gilmore, J.M, Pulavarti, S.V, Baker, D, Szyperski, T. | | Deposit date: | 2016-04-21 | | Release date: | 2016-09-28 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Accurate de novo design of hyperstable constrained peptides.
Nature, 538, 2016
|
|
5TX8
 
 | |
5JG9
 
 | |
7LV6
 
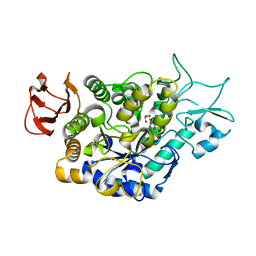 | | The structure of MalL mutant enzyme S536R from Bacillus subtilis | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CALCIUM ION, GLYCEROL, ... | | Authors: | Hamill, C.J, Prentice, E.J, Bahl, C.D, Truebridge, I.S, Arcus, V.L. | | Deposit date: | 2021-02-24 | | Release date: | 2022-03-09 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | Urea binding to guide rational design of mutations that influence enzyme dynamics
To Be Published
|
|
7SOH
 
 | |
