5XLS
 
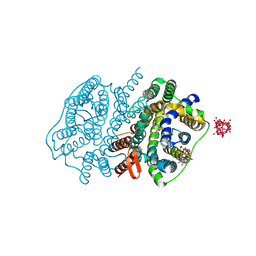 | | Crystal structure of UraA in occluded conformation | | Descriptor: | 12-TUNGSTOPHOSPHATE, URACIL, Uracil permease | | Authors: | Yu, X.Z, Yang, G.H, Yan, C.Y, Yan, N. | | Deposit date: | 2017-05-11 | | Release date: | 2017-07-05 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Dimeric structure of the uracil:proton symporter UraA provides mechanistic insights into the SLC4/23/26 transporters
Cell Res., 27, 2017
|
|
5Z1W
 
 | |
5ZX3
 
 | |
5ZX2
 
 | |
7ERN
 
 | | Crystal structure of D-allulose 3-epimerase with D-fructose from Agrobacterium sp. SUL3 | | Descriptor: | D-fructose, D-tagatose 3-epimerase, MAGNESIUM ION | | Authors: | Zhu, Z.L, Miyakawa, T, Tanokura, M, Lu, F.P, Qin, H.-M. | | Deposit date: | 2021-05-06 | | Release date: | 2022-05-11 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Growth-Coupled Evolutionary Pressure Improving Epimerases for D-Allulose Biosynthesis Using a Biosensor-Assisted In Vivo Selection Platform
Adv Sci, 2024
|
|
7ERO
 
 | | Crystal structure of D-allulose 3-epimerase with D-allulose from Agrobacterium sp. SUL3 | | Descriptor: | D-psicose, D-tagatose 3-epimerase, MAGNESIUM ION | | Authors: | Zhu, Z.L, Miyakawa, T, Tanokura, M, Lu, F.P, Qin, H.-M. | | Deposit date: | 2021-05-06 | | Release date: | 2022-05-11 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.12 Å) | | Cite: | Growth-Coupled Evolutionary Pressure Improving Epimerases for D-Allulose Biosynthesis Using a Biosensor-Assisted In Vivo Selection Platform
Adv Sci, 2024
|
|
7ERM
 
 | | Crystal structure of D-allulose 3-epimerase from Agrobacterium sp. SUL3 | | Descriptor: | D-tagatose 3-epimerase, MAGNESIUM ION, SULFATE ION | | Authors: | Zhu, Z.L, Miyakawa, T, Tanokura, M, Lu, F.P, Qin, H.-M. | | Deposit date: | 2021-05-06 | | Release date: | 2022-05-11 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.32 Å) | | Cite: | Growth-Coupled Evolutionary Pressure Improving Epimerases for D-Allulose Biosynthesis Using a Biosensor-Assisted In Vivo Selection Platform
Adv Sci, 2024
|
|
6J1O
 
 | |
6J46
 
 | | LepI-SAH complex structure | | Descriptor: | O-methyltransferase lepI, S-ADENOSYL-L-HOMOCYSTEINE | | Authors: | Qiu, S, Wei, C. | | Deposit date: | 2019-01-08 | | Release date: | 2019-05-01 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.621 Å) | | Cite: | Deciphering the regulatory and catalytic mechanisms of an unusual SAM-dependent enzyme.
Signal Transduct Target Ther, 4, 2019
|
|
6J24
 
 | | Crystal structure of a SAM-dependent methyltransferase LepI in complex with its substrate | | Descriptor: | (3~{S},4'~{R},4'~{a}~{S},6'~{R},8'~{a}~{S})-4',6'-dimethyl-5-phenyl-spiro[1~{H}-pyridine-3,5'-2,3,4,4~{a},6,8~{a}-hexahydro-1~{H}-naphthalene]-2,4-dione, O-methyltransferase, S-ADENOSYLMETHIONINE | | Authors: | Qiu, S, Wei, C. | | Deposit date: | 2018-12-30 | | Release date: | 2019-05-01 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.244 Å) | | Cite: | Deciphering the regulatory and catalytic mechanisms of an unusual SAM-dependent enzyme.
Signal Transduct Target Ther, 4, 2019
|
|
5XKF
 
 | | Crystal structure of T2R-TTL-MPC6827 complex | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CALCIUM ION, GLYCEROL, ... | | Authors: | Wang, Y, Yang, J, Wang, T, Chen, L. | | Deposit date: | 2017-05-07 | | Release date: | 2018-04-18 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Identification of a powerful and reversible microtubule-inhibitor with efficacy against multidrug-resistant tumors
To Be Published
|
|
5XKE
 
 | | Crystal structure of T2R-TTL-Demecolcine complex | | Descriptor: | (7S)-1,2,3,10-tetramethoxy-7-(methylamino)-6,7-dihydro-5H-benzo[a]heptalen-9-one, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CALCIUM ION, ... | | Authors: | Wang, Y, Yang, J, Wang, T, Chen, L. | | Deposit date: | 2017-05-07 | | Release date: | 2018-07-04 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Identification of a powerful and reversible microtubule-inhibitor with efficacy against multidrug-resistant tumors
To Be Published
|
|
5XIW
 
 | | Crystal structure of T2R-TTL-Colchicine complex | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CALCIUM ION, GLYCEROL, ... | | Authors: | Wang, Y, Yang, J, Wang, T, Chen, L. | | Deposit date: | 2017-04-27 | | Release date: | 2018-04-18 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | The compound millepachine and its derivatives inhibit tubulin polymerization by irreversibly binding to the colchicine-binding site in beta-tubulin.
J. Biol. Chem., 2018
|
|
5XKG
 
 | | Crystal structure of T2R-TTL-CH1 complex | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 4-[(3-azanyl-4-methoxy-phenyl)-methyl-amino]chromen-2-one, CALCIUM ION, ... | | Authors: | Wang, Y, Yang, J, Wang, T, Chen, L. | | Deposit date: | 2017-05-07 | | Release date: | 2018-04-18 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Identification of a powerful and reversible microtubule-inhibitor with efficacy against multidrug-resistant tumors
To Be Published
|
|
5XKH
 
 | | Crystal structure of T2R-TTL-CF1 complex | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 4-[(4-methoxy-3-oxidanyl-phenyl)-methyl-amino]chromen-2-one, CALCIUM ION, ... | | Authors: | Wang, Y, Yang, J, Wang, T, Chen, L. | | Deposit date: | 2017-05-07 | | Release date: | 2018-06-20 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Identification of a powerful and reversible microtubule-inhibitor with efficacy against multidrug-resistant tumors
To Be Published
|
|
5XP3
 
 | | Crystal structure of apo T2R-TTL | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CALCIUM ION, GLYCEROL, ... | | Authors: | Wang, Y, Yang, J, Wang, T, Chen, L. | | Deposit date: | 2017-05-31 | | Release date: | 2017-10-25 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | The compound millepachine and its derivatives inhibit tubulin polymerization by irreversibly binding to the colchicine-binding site in beta-tubulin.
J. Biol. Chem., 2018
|
|
6MU7
 
 | |
6MUG
 
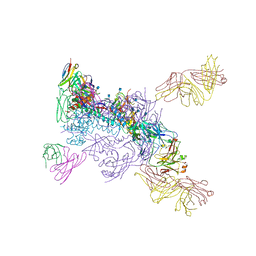 | |
6MTJ
 
 | | Crystal Structure of HIV-1 BG505 SOSIP.664 Prefusion Env Trimer Bound to Small Molecule HIV-1 Entry Inhibitor BMS-378806 in Complex with Human Antibodies 3H109L and 35O22 at 2.9 Angstrom | | Descriptor: | 1-[(2R)-4-(benzenecarbonyl)-2-methylpiperazin-1-yl]-2-(4-methoxy-1H-pyrrolo[2,3-b]pyridin-3-yl)ethane-1,2-dione, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Lai, Y.-T, Kwong, P.D. | | Deposit date: | 2018-10-19 | | Release date: | 2019-01-16 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.336 Å) | | Cite: | Lattice engineering enables definition of molecular features allowing for potent small-molecule inhibition of HIV-1 entry.
Nat Commun, 10, 2019
|
|
6MUF
 
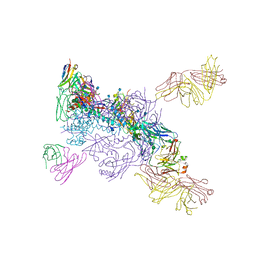 | |
6MTN
 
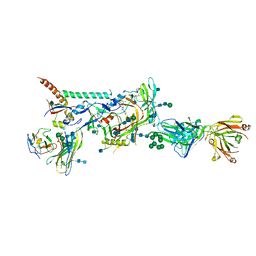 | |
6MU8
 
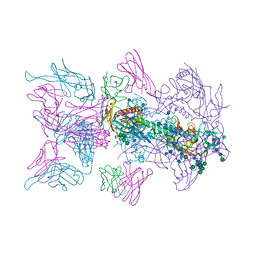 | |
6MU6
 
 | | Crystal Structure of HIV-1 BG505 SOSIP.664 Prefusion Env Trimer Bound to Small Molecule HIV-1 Entry Inhibitor BMS-814508 in Complex with Human Antibodies 3H109L and 35O22 at 3.2 Angstrom | | Descriptor: | (2R)-{1-[{7-[2-({[3-(dimethylamino)propyl](methyl)amino}methyl)-1,3-thiazol-4-yl]-4-methoxy-1H-pyrrolo[2,3-c]pyridin-3-yl}(oxo)acetyl]piperidin-4-yl}(phenyl)acetonitrile, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Lai, Y.-T, Kwong, P.D. | | Deposit date: | 2018-10-22 | | Release date: | 2019-01-16 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.551 Å) | | Cite: | Lattice engineering enables definition of molecular features allowing for potent small-molecule inhibition of HIV-1 entry.
Nat Commun, 10, 2019
|
|
7TRB
 
 | | CRYSTAL STRUCTURE OF FARNESOID X-ACTIVATED RECEPTOR COMPLEXED WITH COMPOUND-32 AKA (1S,3S)-N-({4-[5-(2-FLUOROPR OPAN-2-YL)-1,2,4-OXADIAZOL-3-YL]BICYCLO[2.2.2]OCTAN-1-YL}M ETHYL)-3-HYDROXY-N-[4'-(2-HYDROXYPROPAN-2-YL)-[1,1'-BIPHEN YL]-3-YL]-3-(TRIFLUOROMETHYL)CYCLOBUTANE-1-CARBOXAMIDE | | Descriptor: | (1s,3s)-N-({4-[5-(2-fluoropropan-2-yl)-1,2,4-oxadiazol-3-yl]bicyclo[2.2.2]octan-1-yl}methyl)-3-hydroxy-N-[4'-(2-hydroxypropan-2-yl)[1,1'-biphenyl]-3-yl]-3-(trifluoromethyl)cyclobutane-1-carboxamide, Bile acid receptor, co-activator | | Authors: | Khan, J.A, Ruzanov, M. | | Deposit date: | 2022-01-28 | | Release date: | 2022-06-29 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Discovery of BMS-986339, a Pharmacologically Differentiated Farnesoid X Receptor Agonist for the Treatment of Nonalcoholic Steatohepatitis.
J.Med.Chem., 65, 2022
|
|
7UOQ
 
 | | CRYSTAL STRUCTURE OF HIV-1 INTEGRASE COMPLEXED WITH (2S)-2-(TERT-BUTOXY)-2-(5-{2-[(2-CHLORO-6-M ETHYLPHENYL)METHYL]-1,2,3,4-TETRAHYDROISOQUINOLIN-6-YL}-4- (4,4-DIMETHYLPIPERIDIN-1-YL)-2-METHYLPYRIDIN-3-YL)ACETIC ACID | | Descriptor: | (2S)-tert-butoxy[(5M)-5-{2-[(2-chloro-6-methylphenyl)methyl]-1,2,3,4-tetrahydroisoquinolin-6-yl}-4-(4,4-dimethylpiperidin-1-yl)-2-methylpyridin-3-yl]acetic acid, Integrase, SULFATE ION | | Authors: | Lewis, H.A, Muckelbauer, J.K. | | Deposit date: | 2022-04-13 | | Release date: | 2022-07-06 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.8867 Å) | | Cite: | Discovery and Preclinical Profiling of GSK3839919, a Potent HIV-1 Allosteric Integrase Inhibitor.
Acs Med.Chem.Lett., 13, 2022
|
|
