2H6A
 
 | | Crystal structure of the zinc-beta-lactamase L1 from Stenotrophomonas maltophilia (mono zinc form) | | Descriptor: | Metallo-beta-lactamase L1, SULFATE ION, ZINC ION | | Authors: | Nauton, L, Garau, G, Kahn, R, Dideberg, O. | | Deposit date: | 2006-05-31 | | Release date: | 2007-04-17 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural insights into the design of inhibitors for the L1 metallo-beta-lactamase from Stenotrophomonas maltophilia.
J.Mol.Biol., 375, 2008
|
|
1Q6I
 
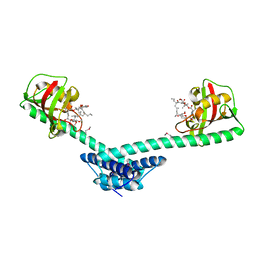 | | Crystal structure of a truncated form of FkpA from Escherichia coli, in complex with immunosuppressant FK506 | | Descriptor: | 8-DEETHYL-8-[BUT-3-ENYL]-ASCOMYCIN, FKBP-type peptidyl-prolyl cis-trans isomerase fkpA | | Authors: | Saul, F.A, Arie, J.-P, Vulliez-le Normand, B, Kahn, R, Betton, J.-M, Bentley, G.A. | | Deposit date: | 2003-08-13 | | Release date: | 2004-01-13 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Structural and functional studies of FkpA from Escherichia coli, a cis/trans peptidyl-prolyl isomerase with chaperone activity.
J.Mol.Biol., 335, 2004
|
|
1Q6U
 
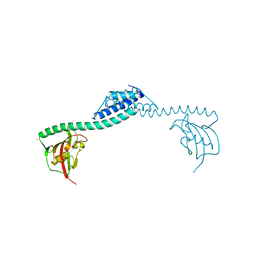 | | Crystal structure of FkpA from Escherichia coli | | Descriptor: | CESIUM ION, FKBP-type peptidyl-prolyl cis-trans isomerase fkpA | | Authors: | Saul, F.A, Arie, J.-P, Vulliez-le Normand, B, Kahn, R, Betton, J.-M, Bentley, G.A. | | Deposit date: | 2003-08-14 | | Release date: | 2004-01-13 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Structural and functional studies of FkpA from Escherichia coli, a cis/trans peptidyl-prolyl isomerase with chaperone activity.
J.Mol.Biol., 335, 2004
|
|
1Q6H
 
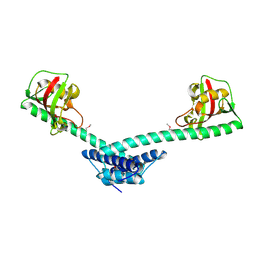 | | Crystal structure of a truncated form of FkpA from Escherichia coli | | Descriptor: | FKBP-type peptidyl-prolyl cis-trans isomerase fkpA | | Authors: | Saul, F.A, Arie, J.-P, Vulliez-le Normand, B, Kahn, R, Betton, J.-M, Bentley, G.A. | | Deposit date: | 2003-08-13 | | Release date: | 2004-01-13 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | Structural and functional studies of FkpA from Escherichia coli, a cis/trans peptidyl-prolyl isomerase with chaperone activity.
J.Mol.Biol., 335, 2004
|
|
2PLO
 
 | |
4BGV
 
 | |
4CL3
 
 | | 1.70 A resolution structure of the malate dehydrogenase from Chloroflexus aurantiacus | | Descriptor: | ACETATE ION, CADMIUM ION, CHLORIDE ION, ... | | Authors: | Talon, R, Madern, D, Girard, E. | | Deposit date: | 2014-01-11 | | Release date: | 2014-02-05 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.699 Å) | | Cite: | An Experimental Point of View on Hydration/Solvation in Halophilic Proteins.
Front.Microbiol., 5, 2014
|
|
4BGU
 
 | | 1.50 A resolution structure of the malate dehydrogenase from Haloferax volcanii | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 3,6,9,12,15,18-HEXAOXAICOSANE-1,20-DIOL, CHLORIDE ION, ... | | Authors: | Talon, R, Madern, D, Girard, E. | | Deposit date: | 2013-03-28 | | Release date: | 2014-04-16 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.487 Å) | | Cite: | Insight Into Structural Evolution of Extremophilic Proteins
To be Published
|
|
1OKJ
 
 | |
3C22
 
 | |
3HW7
 
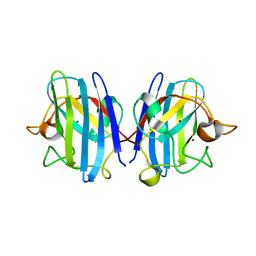 | | High pressure (0.57 GPa) crystal structure of bovine copper, zinc superoxide dismutase at 2.0 angstroms | | Descriptor: | COPPER (I) ION, COPPER (II) ION, Superoxide dismutase [Cu-Zn], ... | | Authors: | Ascone, I, Savino, C. | | Deposit date: | 2009-06-17 | | Release date: | 2010-06-23 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Flexibility of the Cu,Zn superoxide dismutase structure investigated at 0.57 GPa
Acta Crystallogr.,Sect.D, 66, 2010
|
|
2VYU
 
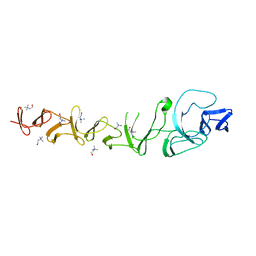 | | CRYSTAL STRUCTURE OF CHOLINE BINDING PROTEIN F FROM STREPTOCOCCUS PNEUMONIAE IN THE PRESENCE OF A PEPTIDOGLYCAN ANALOGUE (TETRASACCHARIDE-PENTAPEPTIDE) | | Descriptor: | CHOLINE BINDING PROTEIN F, CHOLINE ION | | Authors: | Perez-Dorado, I, Molina, R, Hermoso, J.A, Mobashery, S. | | Deposit date: | 2008-07-28 | | Release date: | 2009-02-03 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Crystal Structure of Cbpf, a Bifunctional Choline-Binding Protein and Autolysis Regulator from Streptococcus Pneumoniae.
Embo Rep., 10, 2009
|
|
2TUN
 
 | |
3OJO
 
 | | Derivative structure of the UDP-N-acetyl-mannosamine dehydrogenase Cap5O from S. aureus | | Descriptor: | Cap5O, EUROPIUM ION, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, ... | | Authors: | Nessler, S, Gruszczyk, J, Olivares-Illana, V, Meyer, P, Morera, S. | | Deposit date: | 2010-08-23 | | Release date: | 2011-03-23 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structure Analysis of the Staphylococcus aureus UDP-N-acetyl-mannosamine Dehydrogenase Cap5O Involved in Capsular Polysaccharide Biosynthesis.
J.Biol.Chem., 286, 2011
|
|
3OJL
 
 | | Native structure of the UDP-N-acetyl-mannosamine dehydrogenase Cap5O from Staphylococcus aureus | | Descriptor: | Cap5O, NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | Authors: | Nessler, S, Gruszczyk, J, Olivares-Illana, V, Meyer, P, Morera, S, Grangeasse, C, Fleurie, A. | | Deposit date: | 2010-08-23 | | Release date: | 2011-03-23 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structure Analysis of the Staphylococcus aureus UDP-N-acetyl-mannosamine Dehydrogenase Cap5O Involved in Capsular Polysaccharide Biosynthesis.
J.Biol.Chem., 286, 2011
|
|
5DPX
 
 | | 1,2,4-Triazole-3-thione compounds as inhibitors of L1, di-zinc metallo-beta-lactamases. | | Descriptor: | 5-(2-methylphenyl)-3H-1,2,4-triazole-3-thione, Metallo-beta-lactamase L1 type 3, SULFATE ION, ... | | Authors: | Nauton, L, Garau, G, Khan, R, Dideberg, O. | | Deposit date: | 2015-09-14 | | Release date: | 2017-01-11 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | 1,2,4-Triazole-3-thione Compounds as Inhibitors of Dizinc Metallo-beta-lactamases.
ChemMedChem, 12, 2017
|
|
2KSQ
 
 | | The myristoylated yeast ARF1 in a GTP and bicelle bound conformation | | Descriptor: | ADP-ribosylation factor 1, GUANOSINE-5'-TRIPHOSPHATE, S-[(1-oxyl-2,2,5,5-tetramethyl-2,5-dihydro-1H-pyrrol-3-yl)methyl] methanesulfonothioate | | Authors: | Liu, Y, Kahn, R, Prestegard, J. | | Deposit date: | 2010-01-12 | | Release date: | 2010-07-07 | | Last modified: | 2011-07-27 | | Method: | SOLUTION NMR | | Cite: | Dynamic structure of membrane-anchored Arf*GTP.
Nat.Struct.Mol.Biol., 17, 2010
|
|
1MSB
 
 | |
1DXM
 
 | | Reduced form of the H protein from glycine decarboxylase complex | | Descriptor: | DIHYDROLIPOIC ACID, H PROTEIN | | Authors: | Faure, M, Cohen-Addad, C, Neuburger, M, Douce, R. | | Deposit date: | 2000-01-10 | | Release date: | 2000-07-04 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Interaction between the Lipoamide-Containing H-Protein and the Lipoamide Dehydrogenase (L-Protein) of the Glycine Decarboxylase Multienzyme System. 2. Crystal Structure of H- and L-Proteins
Eur.J.Biochem., 267, 2000
|
|
1DXL
 
 | | Dihydrolipoamide dehydrogenase of glycine decarboxylase from Pisum Sativum | | Descriptor: | DIHYDROLIPOAMIDE DEHYDROGENASE, FLAVIN-ADENINE DINUCLEOTIDE | | Authors: | Faure, M, Cohen-Addad, C, Bourguignon, J, Macherel, D, Neuburger, M, Douce, R. | | Deposit date: | 2000-01-10 | | Release date: | 2000-07-20 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (3.15 Å) | | Cite: | Interaction between the Lipoamide-Containing H-Protein and the Lipoamide Dehydrogenase (L-Protein) of the Glycine Decarboxylase Multienzyme System. 2. Crystal Structure of H- and L-Proteins
Eur.J.Biochem., 267, 2000
|
|
3NEP
 
 | |
1C1M
 
 | | PORCINE ELASTASE UNDER XENON PRESSURE (8 BAR) | | Descriptor: | CALCIUM ION, PROTEIN (PORCINE ELASTASE), SULFATE ION, ... | | Authors: | Prange, T, Schiltz, M, Pernot, L, Colloc'h, N, Longhi, S, Bourguet, W, Fourme, R. | | Deposit date: | 1999-07-22 | | Release date: | 1999-07-28 | | Last modified: | 2018-02-28 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Exploring hydrophobic sites in proteins with xenon or krypton.
Proteins, 30, 1998
|
|
2FUB
 
 | | Crystal structure of urate oxidase at 140 MPa | | Descriptor: | 8-AZAXANTHINE, CYSTEINE, Uricase | | Authors: | Colloc'h, N, Girard, E, Fourme, R. | | Deposit date: | 2006-01-26 | | Release date: | 2006-02-14 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | High pressure macromolecular crystallography: The 140-MPa crystal structure at 2.3 A resolution of urate oxidase, a 135-kDa tetrameric assembly
Biochim.Biophys.Acta, 1764, 2006
|
|
1L9Y
 
 | | FEZ-1-Y228A, A Mutant of the Metallo-beta-lactamase from Legionella gormanii | | Descriptor: | CHLORIDE ION, FEZ-1 b-lactamase, GLYCEROL, ... | | Authors: | Garcia-Saez, I, Mercuri, P.S, Galleni, M, Dideberg, O. | | Deposit date: | 2002-03-27 | | Release date: | 2003-07-01 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | Three-dimensional Structure of FEZ-1, a Monomeric Subclass B3 Metallo-beta-lactamase
from Fluoribacter gormanii, in Native Form and in Complex with -Captopril
J.Mol.Biol., 325, 2003
|
|
7ZQL
 
 | | d(CGCGTTAACGCG)2 B-DODECAMER AT 3100 BARS (310 MPa) | | Descriptor: | DNA (5'-D(*CP*GP*CP*GP*AP*AP*TP*TP*CP*GP*CP*G)-3'), MAGNESIUM ION | | Authors: | Prange, T. | | Deposit date: | 2022-05-01 | | Release date: | 2023-03-08 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Behavior of B- and Z-DNA Crystals under High Hydrostatic Pressure
Crystals, 2022
|
|
