3K7V
 
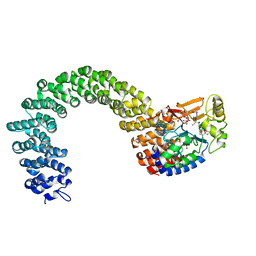 | | Protein phosphatase 2A core complex bound to dinophysistoxin-1 | | Descriptor: | (2R)-3-[(2S,5R,6R,8S)-8-{(1R,2E)-3-[(2R,4a'R,5R,6'S,8'R,8a'S)-6'-{(1S,3S)-3-[(2S,3R,6R,11R)-3,11-dimethyl-1,7-dioxaspiro[5.5]undec-2-yl]-1-hydroxybutyl}-8'-hydroxy-7'-methylideneoctahydro-3H,3'H-spiro[furan-2,2'-pyrano[3,2-b]pyran]-5-yl]-1-methylprop-2-en-1-yl}-5-hydroxy-10-methyl-1,7-dioxaspiro[5.5]undec-10-en-2-yl]-2-hydroxy-2-methylpropanoic acid, MANGANESE (II) ION, SULFATE ION, ... | | Authors: | Jeffrey, P.D, Huhn, J, Shi, Y. | | Deposit date: | 2009-10-13 | | Release date: | 2009-11-03 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | A structural basis for the reduced toxicity of dinophysistoxin-2.
Chem.Res.Toxicol., 22, 2009
|
|
7S96
 
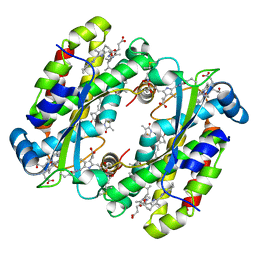 | | Structure of the Light Harvesting Complex PC577 from Hemiselmis pacifica | | Descriptor: | 15,16-DIHYDROBILIVERDIN, PHYCOCYANOBILIN, Phycoerythrin alpha subunit 1, ... | | Authors: | Jeffrey, P.D, Spangler, L.C, Scholes, G.D. | | Deposit date: | 2021-09-20 | | Release date: | 2022-02-09 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Controllable Phycobilin Modification: An Alternative Photoacclimation Response in Cryptophyte Algae.
Acs Cent.Sci., 8, 2022
|
|
7S97
 
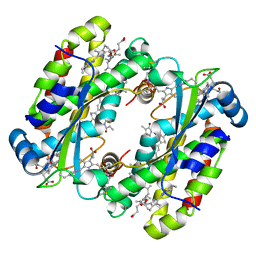 | | Structure of the Photoacclimated Light Harvesting Complex PC577 from Hemiselmis pacifica | | Descriptor: | 15,16-DIHYDROBILIVERDIN, PHYCOCYANOBILIN, Phycoerythrin alpha subunit 1, ... | | Authors: | Jeffrey, P.D, Spangler, L.C, Scholes, G.D. | | Deposit date: | 2021-09-20 | | Release date: | 2022-02-09 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Controllable Phycobilin Modification: An Alternative Photoacclimation Response in Cryptophyte Algae.
Acs Cent.Sci., 8, 2022
|
|
7TLF
 
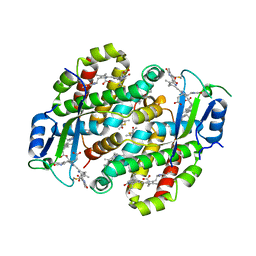 | | Structure of the photoacclimated Light Harvesting Complex PE545 from Proteomonas sulcata | | Descriptor: | 15,16-DIHYDROBILIVERDIN, PHYCOERYTHROBILIN, Phycoerythrin alpha-subunit 1, ... | | Authors: | Jeffrey, P.D, Spangler, L.C, Scholes, G.D. | | Deposit date: | 2022-01-18 | | Release date: | 2022-02-09 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Controllable Phycobilin Modification: An Alternative Photoacclimation Response in Cryptophyte Algae.
Acs Cent.Sci., 8, 2022
|
|
7TJA
 
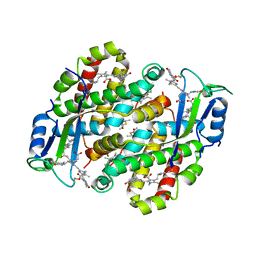 | | Structure of the Light Harvesting Complex PE545 from Proteomonas sulcata | | Descriptor: | 15,16-DIHYDROBILIVERDIN, MAGNESIUM ION, PHYCOERYTHROBILIN, ... | | Authors: | Jeffrey, P.D, Spangler, L.C, Scholes, G.D. | | Deposit date: | 2022-01-15 | | Release date: | 2022-04-20 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | Controllable Phycobilin Modification: An Alternative Photoacclimation Response in Cryptophyte Algae.
Acs Cent.Sci., 8, 2022
|
|
7TZ9
 
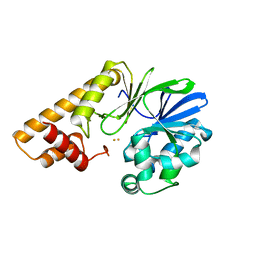 | |
7TZA
 
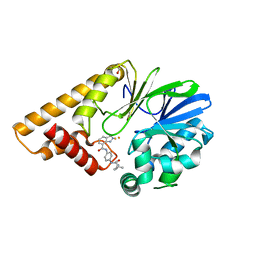 | | Structure of PQS Response Protein PqsE in complex with N-(4-(3-neopentylureido)phenyl)-1H-indazole-7-carboxamide | | Descriptor: | 1,2-ETHANEDIOL, FE (III) ION, N-{4-[(2,2-dimethylpropyl)carbamamido]phenyl}-1H-indazole-7-carboxamide, ... | | Authors: | Jeffrey, P.D, Taylor, I.R, Bassler, B.L. | | Deposit date: | 2022-02-15 | | Release date: | 2022-08-31 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | The PqsE Active Site as a Target for Small Molecule Antimicrobial Agents against Pseudomonas aeruginosa.
Biochemistry, 61, 2022
|
|
7U6G
 
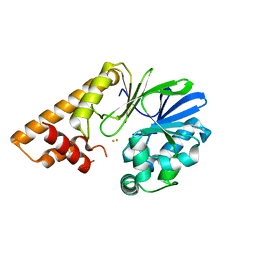 | |
3H11
 
 | |
3H4P
 
 | | Proteasome 20S core particle from Methanocaldococcus jannaschii | | Descriptor: | Proteasome subunit alpha, Proteasome subunit beta | | Authors: | Jeffrey, P.D, Zhang, F, Hu, M, Tian, G, Zhang, P, Finley, D, Shi, Y. | | Deposit date: | 2009-04-20 | | Release date: | 2009-06-09 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (4.1 Å) | | Cite: | Structural Insights into the Regulatory Particle of the Proteasome from Methanocaldococcus jannaschii.
Mol.Cell, 34, 2009
|
|
3H13
 
 | | c-FLIPL protease-like domain | | Descriptor: | CASP8 and FADD-like apoptosis regulator | | Authors: | Jeffrey, P.D, Yu, J.W, Shi, Y. | | Deposit date: | 2009-04-10 | | Release date: | 2009-04-28 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Mechanism of procaspase-8 activation by c-FLIPL.
Proc.Natl.Acad.Sci.USA, 106, 2009
|
|
3H43
 
 | | N-terminal domain of the proteasome-activating nucleotidase of Methanocaldococcus jannaschii | | Descriptor: | Proteasome-activating nucleotidase | | Authors: | Jeffrey, P.D, Zhang, F, Hu, M, Tian, G, Zhang, P, Finley, D, Shi, Y. | | Deposit date: | 2009-04-17 | | Release date: | 2009-06-09 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural Insights into the Regulatory Particle of the Proteasome from Methanocaldococcus jannaschii.
Mol.Cell, 34, 2009
|
|
4U6U
 
 | |
6PRK
 
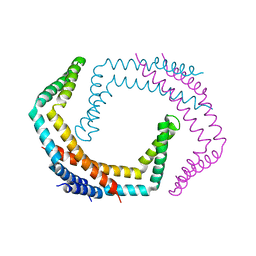 | | X-ray Crystal Structure of Bacillus subtilis RicA in complex with RicF | | Descriptor: | RicA, RicF | | Authors: | Khaja, F.T, Jeffrey, P.D, Neiditch, M.B, Dubnau, D. | | Deposit date: | 2019-07-10 | | Release date: | 2019-10-02 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Structure-Function Studies of the Bacillus subtilis Ric Proteins Identify the Fe-S Cluster-Ligating Residues and Their Roles in Development and RNA Processing.
Mbio, 10, 2019
|
|
5BV0
 
 | |
5BUZ
 
 | |
5BV1
 
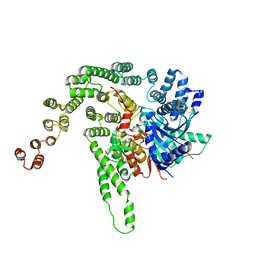 | |
5VAE
 
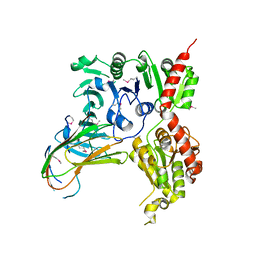 | |
5VAF
 
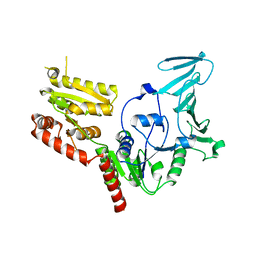 | |
1YY8
 
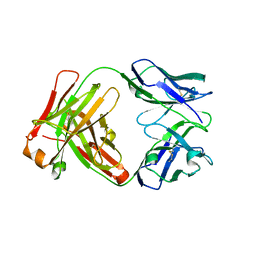 | | Crystal structure of the Fab fragment from the monoclonal antibody cetuximab/Erbitux/IMC-C225 | | Descriptor: | Cetuximab Fab Heavy chain, Cetuximab Fab Light chain | | Authors: | Li, S, Schmitz, K.R, Jeffrey, P.D, Wiltzius, J.J.W, Kussie, P, Ferguson, K.M. | | Deposit date: | 2005-02-24 | | Release date: | 2005-04-26 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural basis for inhibition of the epidermal growth factor receptor by cetuximab
Cancer Cell, 7, 2005
|
|
1YY9
 
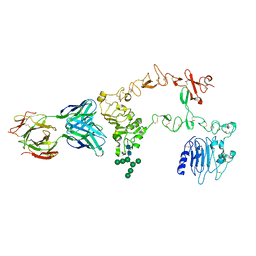 | | Structure of the extracellular domain of the epidermal growth factor receptor in complex with the Fab fragment of cetuximab/Erbitux/IMC-C225 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Cetuximab Fab Heavy chain, ... | | Authors: | Li, S, Schmitz, K.R, Jeffrey, P.D, Wiltzius, J.J.W, Kussie, P, Ferguson, K.M. | | Deposit date: | 2005-02-24 | | Release date: | 2005-04-26 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.605 Å) | | Cite: | Structural basis for inhibition of the epidermal growth factor receptor by cetuximab
Cancer Cell, 7, 2005
|
|
2AYO
 
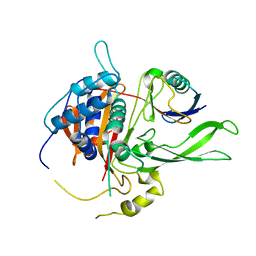 | | Structure of USP14 bound to ubquitin aldehyde | | Descriptor: | Ubiquitin, Ubiquitin carboxyl-terminal hydrolase 14 | | Authors: | Hu, M, Li, P, Jeffrey, P.D, Shi, Y. | | Deposit date: | 2005-09-07 | | Release date: | 2005-10-18 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Structure and mechanisms of the proteasome-associated deubiquitinating enzyme USP14.
Embo J., 24, 2005
|
|
2AYN
 
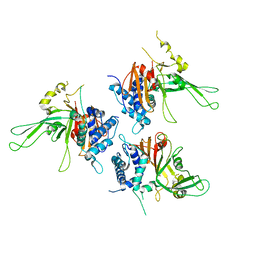 | | Structure of USP14, a proteasome-associated deubiquitinating enzyme | | Descriptor: | Ubiquitin carboxyl-terminal hydrolase 14 | | Authors: | Hu, M, Li, P, Jeffrey, P.D, Shi, Y. | | Deposit date: | 2005-09-07 | | Release date: | 2005-10-18 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Structure and mechanisms of the proteasome-associated deubiquitinating enzyme USP14.
Embo J., 24, 2005
|
|
6CFI
 
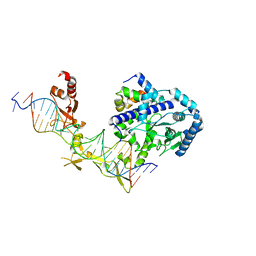 | | Crystal structure of Rad4-Rad23 bound to a 6-4 photoproduct UV lesion | | Descriptor: | DNA (5'-D(*AP*TP*TP*GP*TP*AP*GP*CP*(T64)P*TP*GP*GP*AP*TP*GP*TP*TP*GP*AP*GP*TP*CP*A)-3'), DNA repair protein RAD4, DNA('-D(*TP*TP*GP*AP*CP*TP*CP*AP*AP*CP*AP*TP*CP*CP*AP*AP*AP*GP*CP*TP*AP*CP*AP*A)-'), ... | | Authors: | Min, J, Jeffrey, P.D. | | Deposit date: | 2018-02-15 | | Release date: | 2019-02-27 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (3.36241913 Å) | | Cite: | Structure and mechanism of pyrimidine-pyrimidone (6-4) photoproduct recognition by the Rad4/XPC nucleotide excision repair complex.
Nucleic Acids Res., 47, 2019
|
|
6D0H
 
 | |
