8JNK
 
 | |
8JNR
 
 | |
1JM4
 
 | | NMR Structure of P/CAF Bromodomain in Complex with HIV-1 Tat Peptide | | Descriptor: | HIV-1 Tat Peptide, P300/CBP-associated Factor | | Authors: | Mujtaba, S, He, Y, Zeng, L, Farooq, A, Carlson, J.E, Ott, M, Verdin, E, Zhou, M.-M. | | Deposit date: | 2001-07-17 | | Release date: | 2002-07-17 | | Last modified: | 2023-11-15 | | Method: | SOLUTION NMR | | Cite: | Structural basis of lysine-acetylated HIV-1 Tat recognition by PCAF bromodomain
Mol.Cell, 9, 2002
|
|
3BI3
 
 | |
3BIE
 
 | |
5D9Y
 
 | | Crystal structure of TET2-5fC complex | | Descriptor: | DNA (5'-D(*AP*CP*TP*GP*TP*(5FC)P*GP*AP*AP*GP*CP*T)-3'), DNA (5'-D(*AP*GP*CP*TP*TP*CP*GP*AP*CP*AP*GP*T)-3'), FE (III) ION, ... | | Authors: | Hu, L, Cheng, J, Rao, Q, Li, Z, Li, J, Xu, Y. | | Deposit date: | 2015-08-19 | | Release date: | 2015-11-04 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.971 Å) | | Cite: | Structural insight into substrate preference for TET-mediated oxidation.
Nature, 527, 2015
|
|
5DEU
 
 | | Crystal structure of TET2-5hmC complex | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CHLORIDE ION, DNA (5'-D(*AP*CP*CP*AP*CP*(5HC)P*GP*GP*TP*GP*GP*T)-3'), ... | | Authors: | Hu, L, Cheng, J, Rao, Q, Li, Z, Li, J, Xu, Y. | | Deposit date: | 2015-08-26 | | Release date: | 2015-11-04 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.801 Å) | | Cite: | Structural insight into substrate preference for TET-mediated oxidation.
Nature, 527, 2015
|
|
3H8O
 
 | | Structure determination of DNA methylation lesions N1-meA and N3-meC in duplex DNA using a cross-linked host-guest system | | Descriptor: | 5'-D(*CP*TP*GP*TP*AP*TP*(2YR)P*AP*TP*(MA7)P*GP*CP*G)-3', 5'-D(*TP*CP*GP*CP*TP*AP*TP*AP*AP*TP*AP*CP*A)-3', Alpha-ketoglutarate-dependent dioxygenase alkB homolog 2, ... | | Authors: | Lu, L, Yi, C, Jian, X, Zheng, Q. | | Deposit date: | 2009-04-29 | | Release date: | 2010-03-31 | | Last modified: | 2021-10-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure determination of DNA methylation lesions N1-meA and N3-meC in duplex DNA using a cross-linked protein-DNA system.
Nucleic Acids Res., 38, 2010
|
|
5ZAS
 
 | | Crystal structure of 5-formylcytosine containing decamer dsDNA | | Descriptor: | BICARBONATE ION, DNA (5'-D(*CP*CP*AP*GP*(5FC)P*GP*CP*TP*GP*G)-3') | | Authors: | Fu, T.R, Zhang, L. | | Deposit date: | 2018-02-08 | | Release date: | 2019-02-13 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.56 Å) | | Cite: | Thymine DNA glycosylase recognizes the geometry alteration of minor grooves induced by 5-formylcytosine and 5-carboxylcytosine.
Chem Sci, 10, 2019
|
|
6UIN
 
 | | Role of Beta-hairpin motifs in the DNA duplex opening by the Rad4/XPC nucleotide excision repair complex | | Descriptor: | DNA (5'-D(*AP*TP*TP*GP*TP*AP*GP*NP*NP*NP*NP*GP*GP*AP*TP*GP*TP*CP*GP*AP*GP*TP*CP*A)-3'), DNA (5'-D(*TP*TP*GP*AP*CP*TP*CP*(G47)P*AP*CP*AP*TP*CP*CP*CP*CP*CP*CP*CP*TP*AP*CP*AP*A)-3'), DNA repair protein RAD4, ... | | Authors: | Paul, D, Min, J.-H. | | Deposit date: | 2019-10-01 | | Release date: | 2020-10-14 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (3.348 Å) | | Cite: | Tethering-facilitated DNA 'opening' and complementary roles of beta-hairpin motifs in the Rad4/XPC DNA damage sensor protein
Nucleic Acids Res., 48, 2021
|
|
6UG1
 
 | | Sequence impact in DNA duplex opening by the Rad4/XPC nucleotide excision repair complex | | Descriptor: | DNA (5'-D(*AP*TP*TP*GP*TP*AP*GP*GP*GP*AP*TP*GP*TP*CP*GP*AP*GP*TP*CP*A)-3'), DNA (5'-D(*TP*TP*GP*AP*CP*TP*CP*(G47)P*AP*CP*AP*TP*CP*CP*CP*CP*TP*AP*CP*AP*A)-3'), DNA repair protein RAD4, ... | | Authors: | Paul, D, Min, J.-H. | | Deposit date: | 2019-09-25 | | Release date: | 2021-03-31 | | Last modified: | 2021-09-08 | | Method: | X-RAY DIFFRACTION (2.833 Å) | | Cite: | Impact of DNA sequences on DNA 'opening' by the Rad4/XPC nucleotide excision repair complex.
DNA Repair (Amst), 107, 2021
|
|
4R3H
 
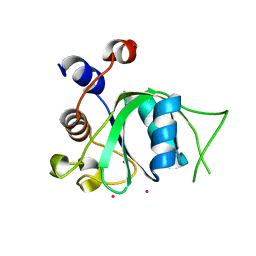 | | The crystal structure of an apo RNA binding protein | | Descriptor: | SULFATE ION, UNKNOWN ATOM OR ION, YTH domain-containing protein 1 | | Authors: | Xu, C, Liu, K, Tempel, W, Li, Y, Bountra, C, Arrowsmith, C.H, Edwards, A.M, Min, J, Structural Genomics Consortium (SGC) | | Deposit date: | 2014-08-15 | | Release date: | 2014-09-17 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural basis for selective binding of m(6)A RNA by the YTHDC1 YTH domain.
Nat.Chem.Biol., 10, 2014
|
|
4R3I
 
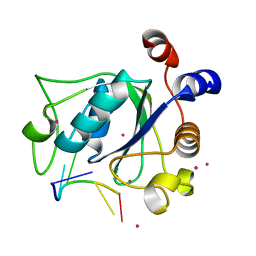 | | The crystal structure of an RNA complex | | Descriptor: | RNA (5'-R(*GP*GP*(6MZ)P*CP*U)-3'), UNKNOWN ATOM OR ION, YTH domain-containing protein 1 | | Authors: | Tempel, W, Xu, C, Liu, K, Li, Y, Bountra, C, Arrowsmith, C.H, Edwards, A.M, Min, J, Structural Genomics Consortium (SGC) | | Deposit date: | 2014-08-15 | | Release date: | 2014-09-17 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural basis for selective binding of m(6)A RNA by the YTHDC1 YTH domain.
Nat.Chem.Biol., 10, 2014
|
|
5YYZ
 
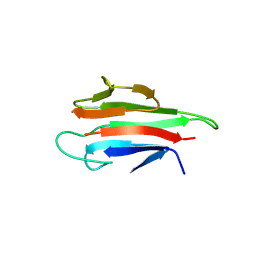 | | Crystal structure of the MEK1 FHA domain in complex with the HOP1 pThr318 peptide. | | Descriptor: | Meiosis-specific protein HOP1, Meiosis-specific serine/threonine-protein kinase MEK1 | | Authors: | Xie, C, Li, F, Jiang, Y, Wu, J, Shi, Y. | | Deposit date: | 2017-12-11 | | Release date: | 2018-10-17 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.798 Å) | | Cite: | Structural insights into the recognition of phosphorylated Hop1 by Mek1
Acta Crystallogr D Struct Biol, 74, 2018
|
|
5YYX
 
 | | Crystal Structure of the MEK1 FHA domain | | Descriptor: | Meiosis-specific serine/threonine-protein kinase MEK1 | | Authors: | Xie, C, Li, F, Jiang, Y, Wu, J, Shi, Y. | | Deposit date: | 2017-12-11 | | Release date: | 2018-10-10 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.684 Å) | | Cite: | Structural insights into the recognition of phosphorylated Hop1 by Mek1
Acta Crystallogr D Struct Biol, 74, 2018
|
|
3VOD
 
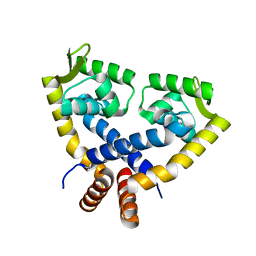 | | Crystal Structure of mutant MarR C80S from E.coli | | Descriptor: | Multiple antibiotic resistance protein marR | | Authors: | Lou, H, Zhu, R, Hao, Z. | | Deposit date: | 2012-01-21 | | Release date: | 2013-03-06 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | The multiple antibiotic resistance regulator MarR is a copper sensor in Escherichia coli.
Nat.Chem.Biol., 10, 2014
|
|
4Y7N
 
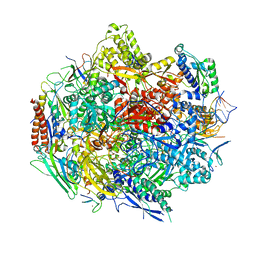 | | The Structure Insight into 5-Carboxycytosine Recognition by RNA Polymerase II during Transcription Elongation. | | Descriptor: | DNA (29-MER), DNA (5'-D(*CP*TP*GP*CP*TP*TP*AP*TP*CP*GP*GP*TP*AP*G)-3'), DNA-directed RNA polymerase II subunit RPB1, ... | | Authors: | Wang, L, Chong, J, Wang, D. | | Deposit date: | 2015-02-15 | | Release date: | 2015-07-15 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Molecular basis for 5-carboxycytosine recognition by RNA polymerase II elongation complex.
Nature, 523, 2015
|
|
4YIR
 
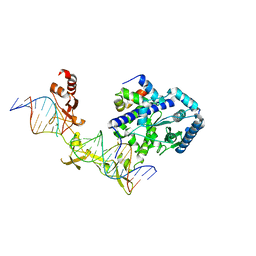 | | Crystal structure of Rad4-Rad23 crosslinked to an undamaged DNA | | Descriptor: | DNA (5'-D(*AP*TP*TP*GP*TP*AP*GP*CP*G*GP*GP*GP*AP*TP*GP*TP*CP*GP*AP*GP*TP*CP*A)-3'), DNA (5'-D(*TP*TP*GP*AP*CP*TP*CP*(G47)P*AP*CP*AP*TP*CP*CP*CP*CP*CP*GP*CP*TP*AP*CP*AP*A)-3'), DNA repair protein RAD4, ... | | Authors: | Min, J.-H, Chen, X, Kim, Y. | | Deposit date: | 2015-03-02 | | Release date: | 2015-03-11 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (3.0501 Å) | | Cite: | Kinetic gating mechanism of DNA damage recognition by Rad4/XPC.
Nat Commun, 6, 2015
|
|
4Y52
 
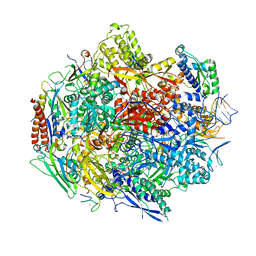 | | Crystal structure of 5-Carboxycytosine Recognition by RNA Polymerase II during Transcription Elongation. | | Descriptor: | DNA (29-MER), DNA (5'-D(*CP*TP*GP*CP*TP*TP*AP*TP*CP*GP*GP*TP*AP*G)-3'), DNA-directed RNA polymerase II subunit RPB1, ... | | Authors: | Wang, L, Chong, J, Wang, D. | | Deposit date: | 2015-02-11 | | Release date: | 2015-07-15 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Molecular basis for 5-carboxycytosine recognition by RNA polymerase II elongation complex.
Nature, 523, 2015
|
|
3QPT
 
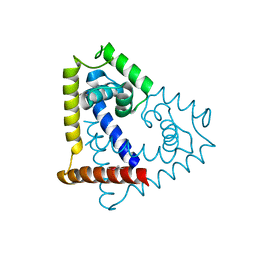 | |
3Q5F
 
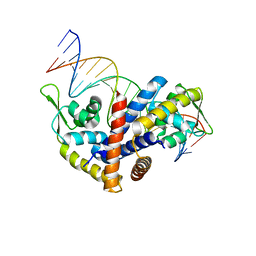 | | Crystal structure of the Salmonella transcriptional regulator SlyA in complex with DNA | | Descriptor: | DNA (5'-D(*AP*AP*TP*AP*AP*CP*TP*TP*AP*GP*CP*AP*AP*GP*CP*TP*AP*AP*TP*TP*AP*TP*A)-3'), DNA (5'-D(*TP*TP*AP*TP*AP*AP*TP*TP*AP*GP*CP*TP*TP*GP*CP*TP*AP*AP*GP*TP*TP*AP*T)-3'), Transcriptional regulator slyA | | Authors: | Dolan, K.T, Duguid, E.M. | | Deposit date: | 2010-12-28 | | Release date: | 2011-05-04 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.96 Å) | | Cite: | Crystal Structures of SlyA Protein, a Master Virulence Regulator of Salmonella, in Free and DNA-bound States.
J.Biol.Chem., 286, 2011
|
|
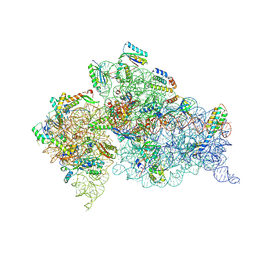
5WNU
 
 | | Crystal Structure of 30S ribosomal subunit from Thermus thermophilus | | Descriptor: | (1R,2S,3S,4R,6R)-4,6-diamino-2-{[3-O-(2,6-diamino-2,6-dideoxy-alpha-L-altropyranosyl)-beta-L-arabinofuranosyl]oxy}-3-hydroxycyclohexyl 2-amino-2-deoxy-alpha-D-allopyranoside, 16S Ribosomal RNA rRNA, 30S ribosomal protein S10, ... | | Authors: | DeMirci, H. | | Deposit date: | 2017-08-01 | | Release date: | 2018-02-21 | | Last modified: | 2020-10-21 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | 2'-O-methylation in mRNA disrupts tRNA decoding during translation elongation.
Nat. Struct. Mol. Biol., 25, 2018
|
|
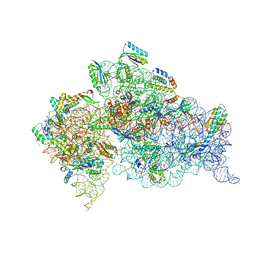
5WNR
 
 | | Crystal Structure of 30S ribosomal subunit from Thermus thermophilus | | Descriptor: | 16S Ribosomal RNA rRNA, 30S ribosomal protein S10, 30S ribosomal protein S11, ... | | Authors: | DeMirci, H. | | Deposit date: | 2017-08-01 | | Release date: | 2018-02-21 | | Last modified: | 2020-10-21 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | 2'-O-methylation in mRNA disrupts tRNA decoding during translation elongation.
Nat. Struct. Mol. Biol., 25, 2018
|
|
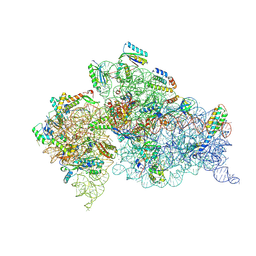
5WNS
 
 | | Crystal Structure of 30S ribosomal subunit from Thermus thermophilus | | Descriptor: | 16S Ribosomal RNA rRNA, 30S ribosomal protein S10, 30S ribosomal protein S11, ... | | Authors: | DeMirci, H. | | Deposit date: | 2017-08-01 | | Release date: | 2018-02-21 | | Last modified: | 2020-10-21 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | 2'-O-methylation in mRNA disrupts tRNA decoding during translation elongation.
Nat. Struct. Mol. Biol., 25, 2018
|
|
5WNQ
 
 | | Crystal Structure of 30S ribosomal subunit from Thermus thermophilus | | Descriptor: | 16S Ribosomal RNA rRNA, 30S ribosomal protein S10, 30S ribosomal protein S11, ... | | Authors: | DeMirci, H. | | Deposit date: | 2017-08-01 | | Release date: | 2018-02-21 | | Last modified: | 2020-10-21 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | 2'-O-methylation in mRNA disrupts tRNA decoding during translation elongation.
Nat. Struct. Mol. Biol., 25, 2018
|
|
