1LTS
 
 | |
1LTT
 
 | |
8AG6
 
 | |
7R2G
 
 | | USP15 D1D2 in catalytically-competent state bound to mitoxantrone stack (isoform 2) | | Descriptor: | 1,4-DIHYDROXY-5,8-BIS({2-[(2-HYDROXYETHYL)AMINO]ETHYL}AMINO)-9,10-ANTHRACENEDIONE, GLYCEROL, Ubiquitin carboxyl-terminal hydrolase 15, ... | | Authors: | Priyanka, A, Sixma, T.K. | | Deposit date: | 2022-02-04 | | Release date: | 2022-06-08 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | Mitoxantrone stacking does not define the active or inactive state of USP15 catalytic domain.
J.Struct.Biol., 214, 2022
|
|
6I5F
 
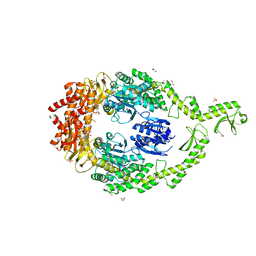 | | Crystal structure of DNA-free E.coli MutS P839E dimer mutant | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, DNA mismatch repair protein MutS, GLYCEROL, ... | | Authors: | Bhairosing-Kok, D, Groothuizen, F.S, Fish, A, Dharadhar, S, Winterwerp, H.H.K, Sixma, T.K. | | Deposit date: | 2018-11-13 | | Release date: | 2019-08-14 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Sharp kinking of a coiled-coil in MutS allows DNA binding and release.
Nucleic Acids Res., 47, 2019
|
|
5FWI
 
 | |
6YN1
 
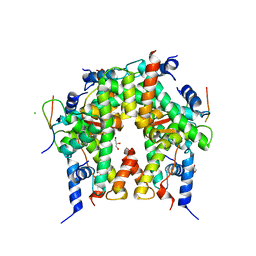 | | Crystal structure of histone chaperone APLF acidic domain bound to the histone H2A-H2B-H3-H4 octamer | | Descriptor: | Aprataxin and PNK-like factor, CHLORIDE ION, GLYCEROL, ... | | Authors: | Corbeski, I, Guo, X, Van Ingen, H, Sixma, T.K. | | Deposit date: | 2020-04-10 | | Release date: | 2021-11-17 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Chaperoning of the histone octamer by the acidic domain of DNA repair factor APLF.
Sci Adv, 8, 2022
|
|
1W7A
 
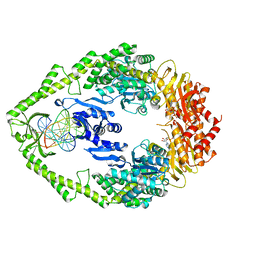 | | ATP bound MutS | | Descriptor: | 5'-D(*AP*GP*CP*TP*GP*CP*CP*AP*GP*GP *CP*AP*CP*CP*AP*GP*TP*GP*TP*CP*AP*GP*CP*GP*TP*CP*CP*TP* AP*T)-3', 5'-D(*AP*TP*AP*GP*GP*AP*CP*GP*CP*TP *GP*AP*CP*AP*CP*TP*GP*GP*TP*GP*CP*TP*TP*GP*GP*CP*AP*GP* CP*T)-3', ADENOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Lamers, M.H, Georgijevic, D, Lebbink, J, Winterwerp, H.H.K, Agianian, B, de Wind, N, Sixma, T.K. | | Deposit date: | 2004-08-31 | | Release date: | 2004-09-10 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.27 Å) | | Cite: | ATP Increases the Affinity between Muts ATPase Domains: Implications for ATP Hydrolysis and Conformational Changes
J.Biol.Chem., 279, 2004
|
|
1WBD
 
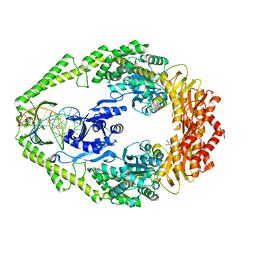 | | Crystal structure of E. coli DNA mismatch repair enzyme MutS, E38Q mutant, in complex with a G.T mismatch | | Descriptor: | 5'-D(*AP*CP*TP*GP*GP*TP*GP*CP*TP*TP *GP*GP*CP*AP*GP*CP*T)-3', 5'-D(*AP*GP*CP*TP*GP*CP*CP*AP*GP*GP *CP*AP*CP*CP*AP*GP*TP*G)-3', ADENOSINE-5'-DIPHOSPHATE, ... | | Authors: | Natrajan, G, Georgijevic, D, Lebbink, J.H.G, Winterwerp, H.H.K, de Wind, N, Sixma, T.K. | | Deposit date: | 2004-10-31 | | Release date: | 2006-01-18 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Dual Role of Muts Glutamate 38 in DNA Mismatch Discrimination and in the Authorization of Repair.
Embo J., 25, 2006
|
|
1WBB
 
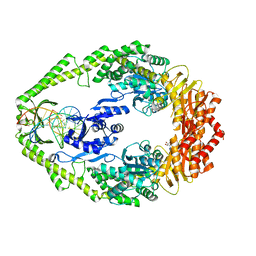 | | Crystal structure of E. coli DNA mismatch repair enzyme MutS, E38A mutant, in complex with a G.T mismatch | | Descriptor: | 5'-D(*AP*CP*TP*GP*GP*TP*GP*CP*TP*TP *GP*GP*CP*AP*GP*CP*T)-3', 5'-D(*AP*GP*CP*TP*GP*CP*CP*AP*GP*GP *CP*AP*CP*CP*AP*GP*TP*G)-3', ADENOSINE-5'-DIPHOSPHATE, ... | | Authors: | Natrajan, G, Georgijevic, D, Lebbink, J.H.G, Winterwerp, H.H.K, de Wind, N, Sixma, T.K. | | Deposit date: | 2004-10-31 | | Release date: | 2006-01-18 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Dual Role of Muts Glutamate 38 in DNA Mismatch Discrimination and in the Authorization of Repair.
Embo J., 25, 2006
|
|
6QLY
 
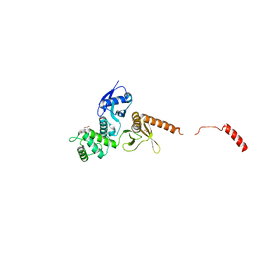 | | IDOL FERM domain | | Descriptor: | 1,2-ETHANEDIOL, E3 ubiquitin-protein ligase MYLIP, SULFATE ION | | Authors: | Martinelli, L, Sixma, T.K. | | Deposit date: | 2019-02-01 | | Release date: | 2020-02-19 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural analysis of the LDL receptor-interacting FERM domain in the E3 ubiquitin ligase IDOL reveals an obscured substrate-binding site.
J.Biol.Chem., 295, 2020
|
|
6QML
 
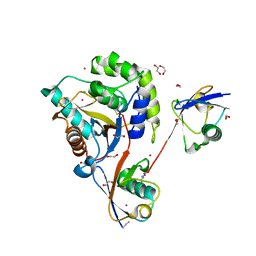 | | UCHL3 in complex with synthetic, K27-linked diubiquitin | | Descriptor: | 1,2-ETHANEDIOL, BROMIDE ION, CHLORIDE ION, ... | | Authors: | Murachelli, A.G, Sixma, T.K. | | Deposit date: | 2019-02-07 | | Release date: | 2020-02-26 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | K27-Linked Diubiquitin Inhibits UCHL3 via an Unusual Kinetic Trap.
Cell Chem Biol, 28, 2021
|
|
4UEM
 
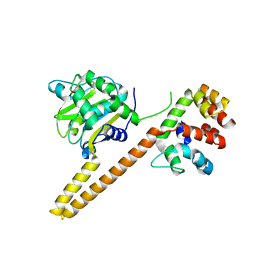 | | UCH-L5 in complex with the RPN13 DEUBAD domain | | Descriptor: | PROTEASOMAL UBIQUITIN RECEPTOR ADRM1, UBIQUITIN CARBOXYL-TERMINAL HYDROLASE ISOZYME L5 | | Authors: | Sahtoe, D.D, Van Dijk, W.J, El Oualid, F, Ekkebus, R, Ovaa, H, Sixma, T.K. | | Deposit date: | 2014-12-18 | | Release date: | 2015-03-04 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.82 Å) | | Cite: | Mechanism of Uch-L5 Activation and Inhibition by Deubad Domains in Rpn13 and Ino80G.
Mol.Cell, 57, 2015
|
|
4UEL
 
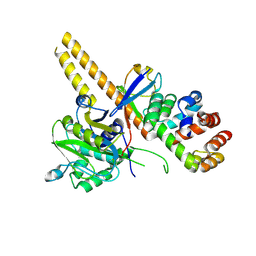 | | UCH-L5 in complex with ubiquitin-propargyl bound to the RPN13 DEUBAD domain | | Descriptor: | POLYUBIQUITIN-B, PROTEASOMAL UBIQUITIN RECEPTOR ADRM1, UBIQUITIN CARBOXYL-TERMINAL HYDROLASE ISOZYME L5 | | Authors: | Sahtoe, D.D, Van Dijk, W.J, El Oualid, F, Ekkebus, R, Ovaa, H, Sixma, T.K. | | Deposit date: | 2014-12-18 | | Release date: | 2015-03-04 | | Last modified: | 2019-04-03 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Mechanism of Uch-L5 Activation and Inhibition by Deubad Domains in Rpn13 and Ino80G.
Mol.Cell, 57, 2015
|
|
4UF5
 
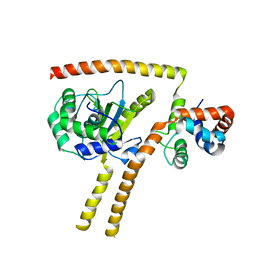 | | Crystal structure of UCH-L5 in complex with inhibitory fragment of INO80G | | Descriptor: | NUCLEAR FACTOR RELATED TO KAPPA-B-BINDING PROTEIN, UBIQUITIN CARBOXYL-TERMINAL HYDROLASE ISOZYME L5 | | Authors: | Sahtoe, D.D, Van Dijk, W.J, El Oualid, F, Ekkebus, R, Ovaa, H, Sixma, T.K. | | Deposit date: | 2014-12-23 | | Release date: | 2015-03-04 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3.7 Å) | | Cite: | Mechanism of Uch-L5 Activation and Inhibition by Deubad Domains in Rpn13 and Ino80G.
Mol.Cell, 57, 2015
|
|
4UF6
 
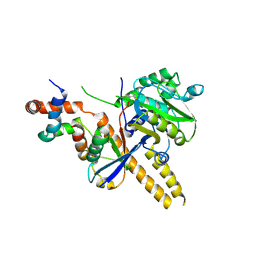 | | UCH-L5 in complex with ubiquitin-propargyl bound to an activating fragment of INO80G | | Descriptor: | NUCLEAR FACTOR RELATED TO KAPPA-B-BINDING PROTEIN, POLYUBIQUITIN-B, UBIQUITIN CARBOXYL-TERMINAL HYDROLASE ISOZYME L5 | | Authors: | Sahtoe, D.D, Van Dijk, W.J, El Oualid, F, Ekkebus, R, Ovaa, H, Sixma, T.K. | | Deposit date: | 2014-12-23 | | Release date: | 2015-03-04 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (3.69 Å) | | Cite: | Mechanism of Uch-L5 Activation and Inhibition by Deubad Domains in Rpn13 and Ino80G.
Mol.Cell, 57, 2015
|
|
3ZLJ
 
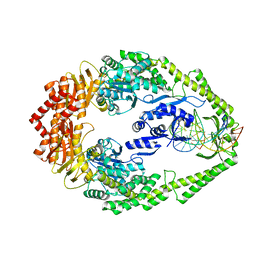 | | CRYSTAL STRUCTURE OF FULL-LENGTH E.COLI DNA MISMATCH REPAIR PROTEIN MUTS D835R MUTANT IN COMPLEX WITH GT MISMATCHED DNA | | Descriptor: | 5'-D(*AP*GP*CP*TP*GP*CP*CP*AP*GP*GP*CP*AP*CP*CP *AP*GP*TP*GP*TP*CP*AP)-3', 5'-D(*TP*GP*AP*CP*AP*CP*TP*GP*GP*TP*GP*CP*TP*TP *GP*GP*CP*AP*GP*CP*TP)-3', DNA MISMATCH REPAIR PROTEIN MUTS | | Authors: | Groothuizen, F.S, Fish, A, Petoukhov, M.V, Reumer, A, Manelyte, L, Winterwerp, H.H.K, Marinus, M.G, Lebbink, J.H.G, Svergun, D.I, Friedhoff, P, Sixma, T.K. | | Deposit date: | 2013-02-01 | | Release date: | 2013-07-17 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Using Stable Muts Dimers and Tetramers to Quantitatively Analyze DNA Mismatch Recognition and Sliding Clamp Formation.
Nucleic Acids Res., 41, 2013
|
|
4AEA
 
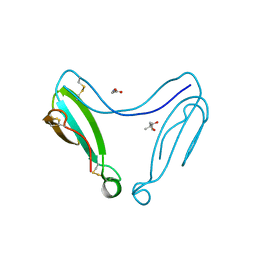 | | Dimeric alpha-cobratoxin X-ray structure: Localization of intermolecular disulfides and possible mode of binding to nicotinic acetylcholine receptors | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, GLYCINE, LONG NEUROTOXIN 1 | | Authors: | Rucktooa, P, Osipov, A.V, Kasheverov, I.E, Filkin, S.Y, Starkov, V.G, Andreeva, T.V, Bertrand, D, Utkin, Y.N, Tsetlin, V.I, Sixma, T.K. | | Deposit date: | 2012-01-09 | | Release date: | 2012-01-25 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | Dimeric Alpha-Cobratoxin X-Ray Structure: Localization of Intermolecular Disulfides and Possible Mode of Binding to Nicotinic Acetylcholine Receptors.
J.Biol.Chem., 287, 2012
|
|
4AFT
 
 | | Aplysia californica AChBP in complex with Varenicline | | Descriptor: | SOLUBLE ACETYLCHOLINE RECEPTOR, VARENICLINE | | Authors: | Rucktooa, P, Haseler, C.A, vanElke, R, Smit, A.B, Gallagher, T, Sixma, T.K. | | Deposit date: | 2012-01-23 | | Release date: | 2012-05-02 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Structural Characterization of Binding Mode of Smoking Cessation Drugs to Nicotinic Acetylcholine Receptors Through Study of Ligand Complexes with Acetylcholine-Binding Protein.
J.Biol.Chem., 287, 2012
|
|
4AYC
 
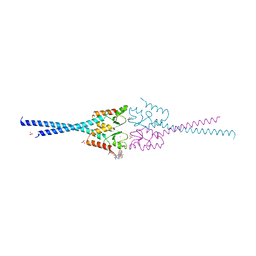 | | RNF8 RING domain structure | | Descriptor: | CHLORIDE ION, E3 UBIQUITIN-PROTEIN LIGASE RNF8, GLYCEROL, ... | | Authors: | Mattiroli, F, Vissers, J.H.A, Van Dijk, W.J, Ikpa, P, Citterio, E, Vermeulen, W, Marteijn, J.A, Sixma, T.K. | | Deposit date: | 2012-06-20 | | Release date: | 2012-09-26 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Rnf168 Ubiquitinates K13-15 on H2A/H2Ax to Drive DNA Damage Signaling
Cell(Cambridge,Mass.), 150, 2012
|
|
5A4P
 
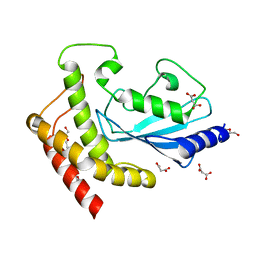 | | Structure of UBE2Z provides functional insight into specificity in the FAT10 conjugation machinery | | Descriptor: | DI(HYDROXYETHYL)ETHER, MALONATE ION, UBIQUITIN-CONJUGATING ENZYME E2 Z | | Authors: | Schelpe, J, Monte, D, Dewitte, F, Sixma, T.K, Rucktooa, P. | | Deposit date: | 2015-06-11 | | Release date: | 2015-11-18 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure of Ube2Z Provides Functional Insight Into Specificity in the Fat10 Conjugation Machinery
J.Biol.Chem., 291, 2016
|
|
5AKD
 
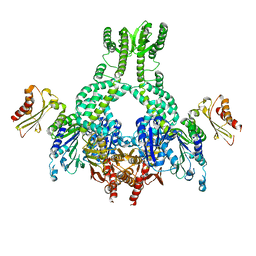 | | MutS in complex with the N-terminal domain of MutL - crystal form 3 | | Descriptor: | DNA MISMATCH REPAIR PROTEIN MUTL, DNA MISMATCH REPAIR PROTEIN MUTS, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER | | Authors: | Groothuizen, F.S, Winkler, I, Cristovao, M, Fish, A, Winterwerp, H.H.K, Reumer, A, Marx, A.D, Hermans, N, Nicholls, R.A, Murshudov, G.N, Lebbink, J.H.G, Friedhoff, P, Sixma, T.K. | | Deposit date: | 2015-03-03 | | Release date: | 2015-07-22 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (7.6 Å) | | Cite: | MutS/MutL crystal structure reveals that the MutS sliding clamp loads MutL onto DNA.
Elife, 4, 2015
|
|
5AKC
 
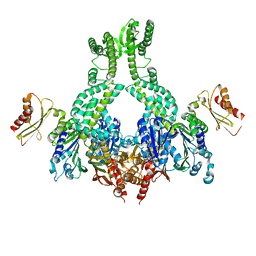 | | MutS in complex with the N-terminal domain of MutL - crystal form 2 | | Descriptor: | DNA MISMATCH REPAIR PROTEIN MUTL, DNA MISMATCH REPAIR PROTEIN MUTS, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER | | Authors: | Groothuizen, F.S, Winkler, I, Cristovao, M, Fish, A, Winterwerp, H.H.K, Reumer, A, Marx, A.D, Hermans, N, Nicholls, R.A, Murshudov, G.N, Lebbink, J.H.G, Friedhoff, P, Sixma, T.K. | | Deposit date: | 2015-03-03 | | Release date: | 2015-07-22 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (6.6 Å) | | Cite: | MutS/MutL crystal structure reveals that the MutS sliding clamp loads MutL onto DNA.
Elife, 4, 2015
|
|
5AKB
 
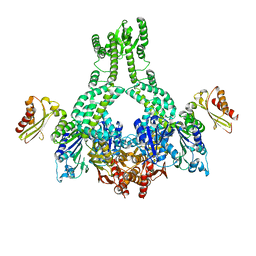 | | MutS in complex with the N-terminal domain of MutL - crystal form 1 | | Descriptor: | DNA MISMATCH REPAIR PROTEIN MUTL, DNA MISMATCH REPAIR PROTEIN MUTS, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER | | Authors: | Groothuizen, F.S, Winkler, I, Cristovao, M, Fish, A, Winterwerp, H.H.K, Reumer, A, Marx, A.D, Hermans, N, Nicholls, R.A, Murshudov, G.N, Lebbink, J.H.G, Friedhoff, P, Sixma, T.K. | | Deposit date: | 2015-03-03 | | Release date: | 2015-07-22 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (4.71 Å) | | Cite: | MutS/MutL crystal structure reveals that the MutS sliding clamp loads MutL onto DNA.
Elife, 4, 2015
|
|
3K0S
 
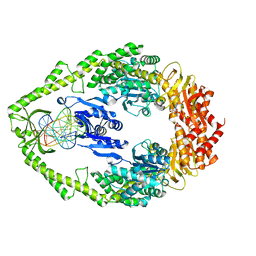 | | Crystal structure of E.coli DNA mismatch repair protein MutS, D693N mutant, in complex with GT mismatched DNA | | Descriptor: | 5'-D(*AP*GP*CP*TP*GP*CP*CP*AP*GP*GP*CP*AP*CP*CP*AP*GP*TP*GP*TP*CP*AP*GP*CP*GP*TP*CP*CP*TP*AP*T)-3', 5'-D(*AP*TP*AP*GP*GP*AP*CP*GP*CP*TP*GP*AP*C*AP*CP*T*GP*GP*TP*GP*CP*TP*TP*GP*GP*CP*AP*GP*CP*T)-3', ADENOSINE-5'-DIPHOSPHATE, ... | | Authors: | Reumer, G.A, Winterwerp, H.H.K, Sixma, T.K. | | Deposit date: | 2009-09-25 | | Release date: | 2010-02-16 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Magnesium coordination controls the molecular switch function of DNA mismatch repair protein MutS.
J.Biol.Chem., 285, 2010
|
|
