6LWL
 
 | | Crystal structure of human NEIL1(R242) bound to duplex DNA containing 2'-fluoro-2'-deoxy-5,6-dihydrouridine | | Descriptor: | DNA (5'-D(*CP*GP*TP*CP*CP*AP*(FDU)P*GP*TP*CP*TP*AP*C)-3'), DNA (5'-D(*TP*AP*GP*AP*CP*CP*TP*GP*GP*AP*CP*GP*G)-3'), Endonuclease 8-like 1, ... | | Authors: | Liu, M.H, Zhang, J, Zhu, C.X, Zhang, X.X, Gao, Y.Q, Yi, C.Q. | | Deposit date: | 2020-02-07 | | Release date: | 2021-06-09 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | DNA repair glycosylase hNEIL1 triages damaged bases via competing interaction modes.
Nat Commun, 12, 2021
|
|
6LWQ
 
 | | Crystal structure of human NEIL1(R242) bound to duplex DNA containing a C:T mismatch | | Descriptor: | DNA (5'-D(*CP*GP*TP*CP*CP*AP*TP*GP*TP*CP*TP*AP*C)-3'), DNA (5'-D(*TP*AP*GP*AP*CP*CP*TP*GP*GP*AP*CP*GP*G)-3'), Endonuclease 8-like 1 | | Authors: | Liu, M.H, Zhang, J, Zhu, C.X, Zhang, X.X, Gao, Y.Q, Yi, C.Q. | | Deposit date: | 2020-02-07 | | Release date: | 2021-06-09 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.89 Å) | | Cite: | DNA repair glycosylase hNEIL1 triages damaged bases via competing interaction modes.
Nat Commun, 12, 2021
|
|
6LWR
 
 | | Crystal structure of human NEIL1(K242) bound to duplex DNA containing a cleaved C:T mismatch | | Descriptor: | DNA (5'-D(*CP*GP*TP*CP*CP*(PDA))-3'), DNA (5'-D(*TP*AP*GP*AP*CP*CP*TP*GP*GP*AP*CP*GP*G)-3'), Endonuclease 8-like 1 | | Authors: | Liu, M.H, Zhang, J, Zhu, C.X, Zhang, X.X, Gao, Y.Q, Yi, C.Q. | | Deposit date: | 2020-02-07 | | Release date: | 2021-06-09 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | DNA repair glycosylase hNEIL1 triages damaged bases via competing interaction modes.
Nat Commun, 12, 2021
|
|
2NO3
 
 | | Novel 4-anilinopyrimidines as potent JNK1 Inhibitors | | Descriptor: | 2-({2-[(3-HYDROXYPHENYL)AMINO]PYRIMIDIN-4-YL}AMINO)BENZAMIDE, C-JUN-AMINO-TERMINAL KINASE-INTERACTING protein 1, Mitogen-activated protein kinase 8, ... | | Authors: | Abad-Zapatero, C. | | Deposit date: | 2006-10-24 | | Release date: | 2007-04-17 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Discovery of a new class of 4-anilinopyrimidines as potent c-Jun N-terminal kinase inhibitors: Synthesis and SAR studies.
Bioorg.Med.Chem.Lett., 17, 2007
|
|
3IWY
 
 | |
3LNJ
 
 | |
6AEX
 
 | | Crystal structure of unoccupied murine uPAR | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Urokinase plasminogen activator surface receptor | | Authors: | Min, L, Huang, M. | | Deposit date: | 2018-08-06 | | Release date: | 2019-07-17 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.393 Å) | | Cite: | Crystal structure of the unoccupied murine urokinase-type plasminogen activator receptor (uPAR) reveals a tightly packed DII-DIII unit.
Febs Lett., 593, 2019
|
|
8I3Q
 
 | |
8FTD
 
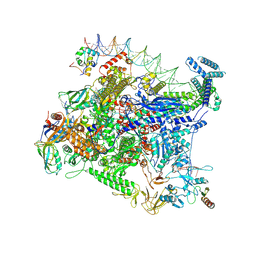 | | Structure of Escherichia coli CedA in complex with transcription initiation complex | | Descriptor: | CHAPSO, Cell division activator CedA, DNA-directed RNA polymerase subunit alpha, ... | | Authors: | Liu, M, Vassyliev, N, Nudler, E. | | Deposit date: | 2023-01-11 | | Release date: | 2024-01-10 | | Last modified: | 2024-01-31 | | Method: | ELECTRON MICROSCOPY (2.76 Å) | | Cite: | General transcription factor from Escherichia coli with a distinct mechanism of action.
Nat.Struct.Mol.Biol., 31, 2024
|
|
8I16
 
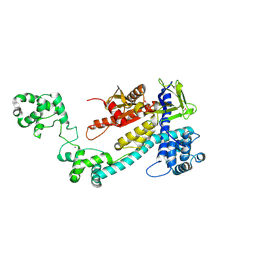 | | Crystal structure of the selenomethionine (SeMet)-derived Cas12g (D513A) mutant | | Descriptor: | Cas12g, ZINC ION | | Authors: | Zhang, B, Chen, J, Ye, Y.M, OuYang, S.Y. | | Deposit date: | 2023-01-12 | | Release date: | 2023-08-30 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.24 Å) | | Cite: | Structural transitions upon guide RNA binding and their importance in Cas12g-mediated RNA cleavage.
Plos Genet., 19, 2023
|
|
7VS2
 
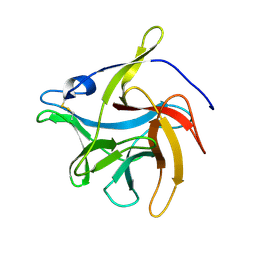 | | secreted fungal effector protein MoErs1 | | Descriptor: | MoErs1 | | Authors: | Wang, F.F, Xing, W.M. | | Deposit date: | 2021-10-25 | | Release date: | 2023-08-02 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Targeting Magnaporthe oryzae effector MoErs1 and host papain-like protease OsRD21 interaction to combat rice blast.
Nat.Plants, 2024
|
|
3C6U
 
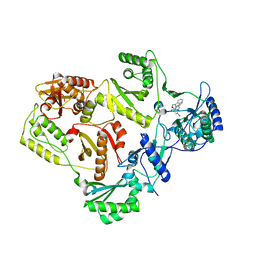 | | Crystal Structure of HIV Reverse Transcriptase in complex with inhibitor 22 | | Descriptor: | 3-chloro-5-[2-chloro-5-(1H-indazol-3-ylmethoxy)phenoxy]benzonitrile, Reverse transcriptase | | Authors: | Yan, Y, Prasad, S. | | Deposit date: | 2008-02-05 | | Release date: | 2008-04-22 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | The design and synthesis of diaryl ether second generation HIV-1 non-nucleoside reverse transcriptase inhibitors (NNRTIs) with enhanced potency versus key clinical mutations.
Bioorg.Med.Chem.Lett., 18, 2008
|
|
6LU7
 
 | | The crystal structure of COVID-19 main protease in complex with an inhibitor N3 | | Descriptor: | 3C-like proteinase, N-[(5-METHYLISOXAZOL-3-YL)CARBONYL]ALANYL-L-VALYL-N~1~-((1R,2Z)-4-(BENZYLOXY)-4-OXO-1-{[(3R)-2-OXOPYRROLIDIN-3-YL]METHYL}BUT-2-ENYL)-L-LEUCINAMIDE | | Authors: | Liu, X, Zhang, B, Jin, Z, Yang, H, Rao, Z. | | Deposit date: | 2020-01-26 | | Release date: | 2020-02-05 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.16 Å) | | Cite: | Structure of Mprofrom SARS-CoV-2 and discovery of its inhibitors.
Nature, 582, 2020
|
|
6L1P
 
 | | Crystal structure of PHF20L1 in complex with Hit 1 | | Descriptor: | 4-(1-methyl-3,6-dihydro-2H-pyridin-4-yl)phenol, GLYCEROL, PHD finger protein 20-like protein 1, ... | | Authors: | Lv, M.Q, Gao, J. | | Deposit date: | 2019-09-29 | | Release date: | 2020-09-23 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.231 Å) | | Cite: | Conformational Selection in Ligand Recognition by the First Tudor Domain of PHF20L1.
J Phys Chem Lett, 11, 2020
|
|
6L1C
 
 | | Crystal Structure Of of PHF20L1 Tudor1 Y24L mutant | | Descriptor: | GLYCEROL, PHD finger protein 20-like protein 1, SULFATE ION | | Authors: | Lv, M.Q, Gao, J. | | Deposit date: | 2019-09-28 | | Release date: | 2020-09-23 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.58 Å) | | Cite: | Conformational Selection in Ligand Recognition by the First Tudor Domain of PHF20L1.
J Phys Chem Lett, 11, 2020
|
|
6L1I
 
 | | Crystal Structure Of of PHF20L1 Tudor1 Y24W/Y29W mutant | | Descriptor: | PHD finger protein 20-like protein 1, SULFATE ION | | Authors: | Lv, M.Q, Gao, J. | | Deposit date: | 2019-09-29 | | Release date: | 2020-09-23 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.849 Å) | | Cite: | Conformational Selection in Ligand Recognition by the First Tudor Domain of PHF20L1.
J Phys Chem Lett, 11, 2020
|
|
6L10
 
 | | PHF20L1 Tudor1 - MES | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, PHD finger protein 20-like protein 1, SULFATE ION | | Authors: | Lv, M.Q, Gao, J. | | Deposit date: | 2019-09-27 | | Release date: | 2020-09-23 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Conformational Selection in Ligand Recognition by the First Tudor Domain of PHF20L1.
J Phys Chem Lett, 11, 2020
|
|
6L1F
 
 | |
4XIR
 
 | | Discovery of novel oxazepine and diazepine carboxamides as two new classes of heat shock protein 90 inhibitors | | Descriptor: | (6S)-2-chloro-8,11,11-trimethyl-9-oxo-6-propyl-6,7,9,10,11,12-hexahydroindolo[2,1-d][1,5]benzoxazepine-3-carboxamide, GLYCEROL, Heat shock protein HSP 90-alpha | | Authors: | Neubert, T, Zuccola, H.J. | | Deposit date: | 2015-01-07 | | Release date: | 2015-03-04 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Discovery of novel oxazepine and diazepine carboxamides as two new classes of heat shock protein 90 inhibitors.
Bioorg.Med.Chem.Lett., 25, 2015
|
|
4XIP
 
 | | Discovery of novel oxazepine and diazepine carboxamides as two new classes of heat shock protein 90 inhibitors | | Descriptor: | 4-(3,6,6-trimethyl-4-oxo-4,5,6,7-tetrahydro-1H-indol-1-yl)benzamide, Heat shock protein HSP 90-alpha | | Authors: | Zuccola, H.J, Neubert, T. | | Deposit date: | 2015-01-07 | | Release date: | 2015-03-04 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Discovery of novel oxazepine and diazepine carboxamides as two new classes of heat shock protein 90 inhibitors.
Bioorg.Med.Chem.Lett., 25, 2015
|
|
4XIQ
 
 | | Discovery of novel oxazepine and diazepine carboxamides as two new classes of heat shock protein 90 inhibitors | | Descriptor: | 8,11,11-trimethyl-9-oxo-6,7,9,10,11,12-hexahydroindolo[2,1-d][1,5]benzoxazepine-3-carboxamide, GLYCEROL, Heat shock protein HSP 90-alpha | | Authors: | Neubert, T, Zuccola, H.J. | | Deposit date: | 2015-01-07 | | Release date: | 2015-03-04 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.84 Å) | | Cite: | Discovery of novel oxazepine and diazepine carboxamides as two new classes of heat shock protein 90 inhibitors.
Bioorg.Med.Chem.Lett., 25, 2015
|
|
4XIT
 
 | | Discovery of novel oxazepine and diazepine carboxamides as two new classes of heat shock protein 90 inhibitors | | Descriptor: | 2-chloro-5-(furan-2-ylmethyl)-8,11,11-trimethyl-9-oxo-6,7,9,10,11,12-hexahydro-5H-indolo[1,2-a][1,5]benzodiazepine-3-carboxamide, GLYCEROL, Heat shock protein HSP 90-alpha | | Authors: | Zuccola, H.J, Neubert, T. | | Deposit date: | 2015-01-07 | | Release date: | 2015-03-04 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | Discovery of novel oxazepine and diazepine carboxamides as two new classes of heat shock protein 90 inhibitors.
Bioorg.Med.Chem.Lett., 25, 2015
|
|
4XUA
 
 | | Crystal Structure of the bromodomain of human BAZ2B in complex with E11919 BAZ2-ICR analogue | | Descriptor: | 1,2-ETHANEDIOL, 4-{1-[2-(4-methyl-1H-1,2,3-triazol-1-yl)ethyl]-4-phenyl-1H-imidazol-5-yl}benzonitrile, Bromodomain adjacent to zinc finger domain protein 2B | | Authors: | Chaikuad, A, Felletar, I, von Delft, F, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2015-01-25 | | Release date: | 2015-03-11 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structure Enabled Design of BAZ2-ICR, A Chemical Probe Targeting the Bromodomains of BAZ2A and BAZ2B.
J.Med.Chem., 58, 2015
|
|
6X4X
 
 | | B24Y DKP insulin | | Descriptor: | Insulin, Insulin chain A | | Authors: | Weiss, M.A, Yang, Y. | | Deposit date: | 2020-05-24 | | Release date: | 2020-08-05 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Evolution of insulin at the edge of foldability and its medical implications.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
8YXR
 
 | |
