2FLA
 
 | | Structure of thereduced HiPIP from thermochromatium tepidum at 0.95 angstrom resolution | | Descriptor: | GLYCEROL, High-potential iron-sulfur protein, IRON/SULFUR CLUSTER, ... | | Authors: | Hunsicker-Wang, L.M, Stout, C.D, Fee, J.A. | | Deposit date: | 2006-01-05 | | Release date: | 2006-12-05 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (0.95 Å) | | Cite: | Geometric factors determine, in part, the electronic state of the 4Fe-4S cluster of HiPIP: a geometric, crystallographic, and theoretical study.
To be Published
|
|
5WB6
 
 | | FACTOR XIA IN COMPLEX WITH THE INHIBITOR methyl [(11S)-11-({(2E)-3-[5-chloro-2-(1H-tetrazol-1-yl)phenyl]prop-2-enoyl}amino)-6-fluoro-2-oxo-1,3,4,10,11,13-hexahydro-2H-5,9:15,12-di(azeno)-1,13-benzodiazacycloheptadecin-18-yl]carbamate | | Descriptor: | 1,2-ETHANEDIOL, Coagulation factor XI, SULFATE ION, ... | | Authors: | Sheriff, S. | | Deposit date: | 2017-06-28 | | Release date: | 2017-08-02 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Macrocyclic factor XIa inhibitors.
Bioorg. Med. Chem. Lett., 27, 2017
|
|
4N6Z
 
 | | Pim1 Complexed with a pyridylcarboxamide | | Descriptor: | 3-amino-N-{4-[(3S)-3-aminopiperidin-1-yl]pyridin-3-yl}pyrazine-2-carboxamide, GLYCEROL, Serine/threonine-protein kinase pim-1 | | Authors: | Bellamacina, C.R, Le, V, Shu, W, Burger, M.T, Bussiere, D. | | Deposit date: | 2013-10-14 | | Release date: | 2013-11-06 | | Last modified: | 2014-07-02 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure Guided Optimization, in Vitro Activity, and in Vivo Activity of Pan-PIM Kinase Inhibitors.
ACS Med Chem Lett, 4, 2013
|
|
4Y8Z
 
 | |
4Y8Y
 
 | |
4Y8X
 
 | |
5BVD
 
 | | Tetrahydropyrrolo-diazepenones as inhibitors of ERK2 kinase | | Descriptor: | 2-[(1S)-1-(3-chlorophenyl)-2-hydroxyethyl]-7-[2-(tetrahydro-2H-pyran-4-ylamino)pyrimidin-4-yl]-3,4-dihydropyrrolo[1,2-a]pyrazin-1(2H)-one, Mitogen-activated protein kinase 1, SULFATE ION | | Authors: | Ma, X, Steven, S. | | Deposit date: | 2015-06-05 | | Release date: | 2015-09-09 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Tetrahydropyrrolo-diazepenones as inhibitors of ERK2 kinase.
Bioorg.Med.Chem.Lett., 25, 2015
|
|
5BVE
 
 | | Tetrahydropyrrolo-diazepenones as inhibitors of ERK2 kinase | | Descriptor: | 2-[(1S)-1-(3-chlorophenyl)-2-hydroxyethyl]-8-[2-(tetrahydro-2H-pyran-4-ylamino)pyrimidin-4-yl]-2,3,4,5-tetrahydro-1H-pyrrolo[1,2-a][1,4]diazepin-1-one, Mitogen-activated protein kinase 1, SULFATE ION | | Authors: | Ma, X, Steven, S. | | Deposit date: | 2015-06-05 | | Release date: | 2015-09-09 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Tetrahydropyrrolo-diazepenones as inhibitors of ERK2 kinase.
Bioorg.Med.Chem.Lett., 25, 2015
|
|
3CEN
 
 | |
5DWR
 
 | | Identification of N-(4-((1R,3S,5S)-3-amino-5-methylcyclohexyl)pyridin-3-yl)-6-(2,6-difluorophenyl)-5-fluoropicolinamide (PIM447), a Potent and Selective Proviral Insertion Site of Moloney Murine Leukemia (PIM) 1,2 and 3 Kinase Inhibitor in Clinical Trials for Hematological Malignancies | | Descriptor: | N-{4-[(1R,3S,5S)-3-amino-5-methylcyclohexyl]pyridin-3-yl}-6-(2,6-difluorophenyl)-5-fluoropyridine-2-carboxamide, Serine/threonine-protein kinase pim-1 | | Authors: | Bellamacina, C, Bussiere, D, Burger, M. | | Deposit date: | 2015-09-22 | | Release date: | 2015-11-11 | | Last modified: | 2015-11-25 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Identification of N-(4-((1R,3S,5S)-3-Amino-5-methylcyclohexyl)pyridin-3-yl)-6-(2,6-difluorophenyl)-5-fluoropicolinamide (PIM447), a Potent and Selective Proviral Insertion Site of Moloney Murine Leukemia (PIM) 1, 2, and 3 Kinase Inhibitor in Clinical Trials for Hematological Malignancies.
J.Med.Chem., 58, 2015
|
|
5HD8
 
 | |
5IIS
 
 | | Design, synthesis and structure activity relationship of potent pan-PIM kinase inhibitors derived from the pyridyl-amide scaffold | | Descriptor: | 3-amino-N-(2'-amino-6'-methyl[4,4'-bipyridin]-3-yl)-6-(2-fluorophenyl)pyridine-2-carboxamide, DI(HYDROXYETHYL)ETHER, Serine/threonine-protein kinase pim-1 | | Authors: | Bellamacina, C, Bussiere, D, Burger, M. | | Deposit date: | 2016-03-01 | | Release date: | 2016-04-06 | | Last modified: | 2016-05-04 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Design, synthesis and structure activity relationship of potent pan-PIM kinase inhibitors derived from the pyridyl carboxamide scaffold.
Bioorg.Med.Chem.Lett., 26, 2016
|
|
5BVF
 
 | | Tetrahydropyrrolo-diazepenones as inhibitors of ERK2 kinase | | Descriptor: | 2-[(1S)-1-(3-chlorophenyl)-2-hydroxyethyl]-8-(2-{[(1S,3R)-3-hydroxycyclopentyl]amino}pyrimidin-4-yl)-2,3,4,5-tetrahydro-1H-pyrrolo[1,2-a][1,4]diazepin-1-one, Mitogen-activated protein kinase 1, SULFATE ION | | Authors: | Ma, X, Steven, S. | | Deposit date: | 2015-06-05 | | Release date: | 2015-09-09 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Tetrahydropyrrolo-diazepenones as inhibitors of ERK2 kinase.
Bioorg.Med.Chem.Lett., 25, 2015
|
|
4KZ0
 
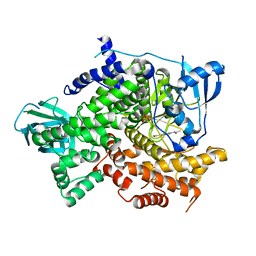 | | Structure of PI3K gamma with Imidazopyridine inhibitors | | Descriptor: | Phosphatidylinositol 4,5-bisphosphate 3-kinase catalytic subunit gamma isoform, SULFATE ION, methyl 2-(acetylamino)-1,3-benzothiazole-6-carboxylate | | Authors: | Knapp, M.S, Elling, R.A. | | Deposit date: | 2013-05-29 | | Release date: | 2013-07-17 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.87 Å) | | Cite: | Structure guided optimization of a fragment hit to imidazopyridine inhibitors of PI3K.
Bioorg.Med.Chem.Lett., 23, 2013
|
|
4KZC
 
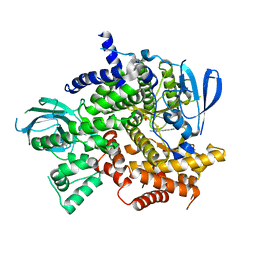 | | Structure of PI3K gamma with Imidazopyridine inhibitors | | Descriptor: | N-{6-[6-amino-5-(trifluoromethyl)pyridin-3-yl]imidazo[1,2-a]pyridin-2-yl}acetamide, Phosphatidylinositol 4,5-bisphosphate 3-kinase catalytic subunit gamma isoform, SULFATE ION | | Authors: | Knapp, M.S, Elling, E.A. | | Deposit date: | 2013-05-29 | | Release date: | 2013-07-17 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (3.25 Å) | | Cite: | Structure guided optimization of a fragment hit to imidazopyridine inhibitors of PI3K.
Bioorg.Med.Chem.Lett., 23, 2013
|
|
3T9I
 
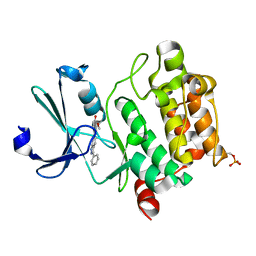 | | Pim1 complexed with a novel 3,6-disubstituted indole at 2.6 Ang Resolution | | Descriptor: | 2-methoxy-4-(3-phenyl-2H-pyrazolo[3,4-b]pyridin-6-yl)phenol, Proto-oncogene serine/threonine-protein kinase pim-1 | | Authors: | Bellamacina, C, Shu, W, Le, V, Nishiguchi, G, Bussiere, D. | | Deposit date: | 2011-08-02 | | Release date: | 2011-10-12 | | Last modified: | 2011-11-02 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Discovery of novel 3,5-disubstituted indole derivatives as potent inhibitors of Pim-1, Pim-2, and Pim-3 protein kinases.
Bioorg.Med.Chem.Lett., 21, 2011
|
|
7WVQ
 
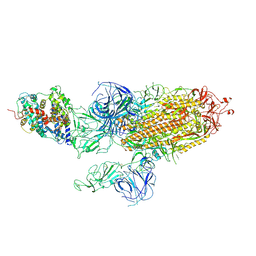 | |
7WVP
 
 | |
6KRE
 
 | | TRiC at 0.05 mM ADP-AlFx, Conformation 2, 0.05-C2 | | Descriptor: | T-complex protein 1 subunit alpha, T-complex protein 1 subunit beta, T-complex protein 1 subunit delta, ... | | Authors: | Jin, M, Cong, Y. | | Deposit date: | 2019-08-21 | | Release date: | 2019-09-18 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (4.45 Å) | | Cite: | An ensemble of cryo-EM structures of TRiC reveal its conformational landscape and subunit specificity.
Proc.Natl.Acad.Sci.USA, 116, 2019
|
|
6KRD
 
 | | TRiC at 0.05 mM ADP-AlFx, Conformation 4, 0.05-C4 | | Descriptor: | T-complex protein 1 subunit alpha, T-complex protein 1 subunit beta, T-complex protein 1 subunit delta, ... | | Authors: | Jin, M, Cong, Y. | | Deposit date: | 2019-08-21 | | Release date: | 2019-09-18 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (4.38 Å) | | Cite: | An ensemble of cryo-EM structures of TRiC reveal its conformational landscape and subunit specificity.
Proc.Natl.Acad.Sci.USA, 116, 2019
|
|
6KS8
 
 | | TRiC at 0.1 mM ADP-AlFx, Conformation 4, 0.1-C4 | | Descriptor: | T-complex protein 1 subunit alpha, T-complex protein 1 subunit beta, T-complex protein 1 subunit delta, ... | | Authors: | Jin, M, Cong, Y. | | Deposit date: | 2019-08-23 | | Release date: | 2019-09-18 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (4.69 Å) | | Cite: | An ensemble of cryo-EM structures of TRiC reveal its conformational landscape and subunit specificity.
Proc.Natl.Acad.Sci.USA, 116, 2019
|
|
6KS7
 
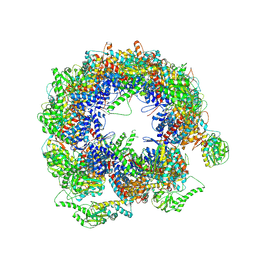 | | TRiC at 0.1 mM ADP-AlFx, Conformation 1, 0.1-C1 | | Descriptor: | T-complex protein 1 subunit alpha, T-complex protein 1 subunit beta, T-complex protein 1 subunit delta, ... | | Authors: | Jin, M, Cong, Y. | | Deposit date: | 2019-08-23 | | Release date: | 2019-09-18 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (4.62 Å) | | Cite: | An ensemble of cryo-EM structures of TRiC reveal its conformational landscape and subunit specificity.
Proc.Natl.Acad.Sci.USA, 116, 2019
|
|
7C8V
 
 | | Structure of sybody SR4 in complex with the SARS-CoV-2 S Receptor Binding domain (RBD) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, GLYCEROL, Spike protein S1, ... | | Authors: | Li, T, Yao, H, Cai, H, Qin, W, Li, D. | | Deposit date: | 2020-06-03 | | Release date: | 2020-06-24 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | A synthetic nanobody targeting RBD protects hamsters from SARS-CoV-2 infection.
Nat Commun, 12, 2021
|
|
7CAN
 
 | | Structure of sybody MR17-K99Y in complex with the SARS-CoV-2 S Receptor-binding domain (RBD) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, GLYCEROL, Spike protein S1, ... | | Authors: | Li, T, Yao, H, Cai, H, Qin, W, Li, D. | | Deposit date: | 2020-06-09 | | Release date: | 2020-06-24 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.94 Å) | | Cite: | A synthetic nanobody targeting RBD protects hamsters from SARS-CoV-2 infection.
Nat Commun, 12, 2021
|
|
7C8W
 
 | | Structure of sybody MR17 in complex with the SARS-CoV-2 S receptor-binding domain (RBD) | | Descriptor: | GLYCEROL, Spike protein S1, Synthetic nanobody MR17, ... | | Authors: | Li, T, Cai, H, Yao, H, Qin, W, Li, D. | | Deposit date: | 2020-06-03 | | Release date: | 2020-06-24 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.77 Å) | | Cite: | A synthetic nanobody targeting RBD protects hamsters from SARS-CoV-2 infection.
Nat Commun, 12, 2021
|
|
