7YBK
 
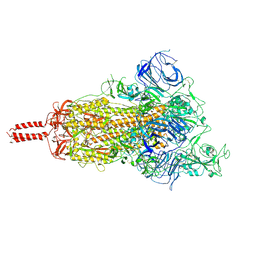 | | SARS-CoV-2 B.1.620 variant spike (open state) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Wang, X, Fu, W. | | Deposit date: | 2022-06-29 | | Release date: | 2023-09-06 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Structures of SARS-CoV-2 spike protein alert noteworthy sites for the potential approaching variants.
Virol Sin, 37, 2022
|
|
6IRR
 
 | | Solution structure of DISC1/ATF4 complex | | Descriptor: | Disrupted in schizophrenia 1 homolog,Cyclic AMP-dependent transcription factor ATF-4 | | Authors: | Ye, F, Yu, C, Zhang, M. | | Deposit date: | 2018-11-14 | | Release date: | 2019-09-25 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structural interaction between DISC1 and ATF4 underlying transcriptional and synaptic dysregulation in an iPSC model of mental disorders.
Mol. Psychiatry, 2019
|
|
7WDH
 
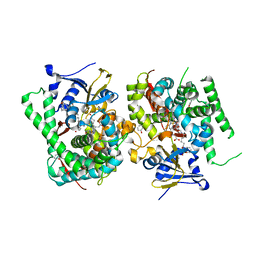 | | Crystal structure of the P450 BM3 heme domain mutant F87A in complex with N-imidazolyl-hexanoyl-L-phenylalanine, phenol and hydroxylamine | | Descriptor: | (2S)-2-(6-imidazol-1-ylhexanoylamino)-3-phenyl-propanoic acid, Bifunctional cytochrome P450/NADPH--P450 reductase, HYDROXYAMINE, ... | | Authors: | Jiang, Y, Dong, S, Feng, Y, Cong, Z. | | Deposit date: | 2021-12-21 | | Release date: | 2022-12-28 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.68 Å) | | Cite: | Engineering Cytochrome P450BM3 Enzymes for Direct Nitration of Unsaturated Hydrocarbons.
Angew.Chem.Int.Ed.Engl., 62, 2023
|
|
7WDG
 
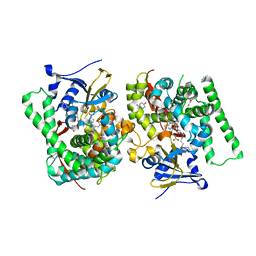 | | Crystal structure of the P450 BM3 heme domain mutant F87L in complex with N-imidazolyl-hexanoyl-L-phenylalanine, phenol and hydroxylamine | | Descriptor: | (2S)-2-(6-imidazol-1-ylhexanoylamino)-3-phenyl-propanoic acid, Bifunctional cytochrome P450/NADPH--P450 reductase, HYDROXYAMINE, ... | | Authors: | Jiang, Y, Dong, S, Feng, Y, Cong, Z. | | Deposit date: | 2021-12-21 | | Release date: | 2022-12-28 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.07 Å) | | Cite: | Engineering Cytochrome P450BM3 Enzymes for Direct Nitration of Unsaturated Hydrocarbons.
Angew.Chem.Int.Ed.Engl., 62, 2023
|
|
7EZP
 
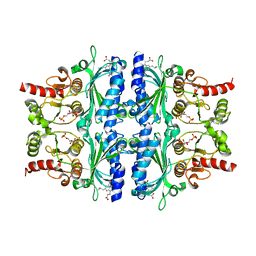 | | Indole-2-carboxylic acid derivatives as allosteric inhibitors of fructose-1,6-bisphosphatase | | Descriptor: | 1,6-di-O-phosphono-beta-D-fructofuranose, 3-(3-hydroxy-3-oxopropyl)-5-(2-methylpropyl)-7-nitro-1H-indole-2-carboxylic acid, Fructose-1,6-bisphosphatase 1 | | Authors: | Wang, X.Y, Zhou, J, Xu, B.L. | | Deposit date: | 2021-06-01 | | Release date: | 2022-06-01 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Discovery of Novel Indole Derivatives as Fructose-1,6-bisphosphatase Inhibitors and X-ray Cocrystal Structures Analysis.
Acs Med.Chem.Lett., 13, 2022
|
|
7EZR
 
 | | Indole-2-carboxylic acid derivatives as allosteric inhibitors of fructose-1,6-bisphosphatase | | Descriptor: | 1,6-di-O-phosphono-beta-D-fructofuranose, 5-ethyl-7-nitro-3-[3-oxidanylidene-3-(thiophen-2-ylsulfonylamino)propyl]-1H-indole-2-carboxylic acid, Fructose-1,6-bisphosphatase 1 | | Authors: | Wang, X.Y, Zhou, J, Xu, B.L. | | Deposit date: | 2021-06-01 | | Release date: | 2022-06-01 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3.27 Å) | | Cite: | Discovery of Novel Indole Derivatives as Fructose-1,6-bisphosphatase Inhibitors and X-ray Cocrystal Structures Analysis.
Acs Med.Chem.Lett., 13, 2022
|
|
7EZF
 
 | | Indole-2-carboxylic acid derivatives as allosteric inhibitors of fructose-1,6-bisphosphatase | | Descriptor: | 1,6-di-O-phosphono-beta-D-fructofuranose, 7-chloranyl-5-ethyl-3-(3-hydroxy-3-oxopropyl)-1H-indole-2-carboxylic acid, Fructose-1,6-bisphosphatase 1 | | Authors: | Wang, X.Y, Zhou, J, Xu, B.L. | | Deposit date: | 2021-06-01 | | Release date: | 2022-06-01 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.76 Å) | | Cite: | Discovery of Novel Indole Derivatives as Fructose-1,6-bisphosphatase Inhibitors and X-ray Cocrystal Structures Analysis.
Acs Med.Chem.Lett., 13, 2022
|
|
7VHH
 
 | | Delta variant of SARS-CoV-2 Spike protein | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein | | Authors: | Liu, P. | | Deposit date: | 2021-09-22 | | Release date: | 2022-12-28 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Structure of delta variant SARS-CoV-2 Spike protein
To Be Published
|
|
5JNX
 
 | | The 6.6 A cryo-EM structure of the full-length human NPC1 in complex with the cleaved glycoprotein of Ebola virus | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Envelope glycoprotein, ... | | Authors: | Gong, X, Qian, H.W, Zhou, X.H, Wu, J.P, Wan, T, Shi, Y, Gao, F, Zhou, Q, Yan, N. | | Deposit date: | 2016-05-01 | | Release date: | 2016-06-15 | | Last modified: | 2020-08-05 | | Method: | ELECTRON MICROSCOPY (6.56 Å) | | Cite: | Structural Insights into the Niemann-Pick C1 (NPC1)-Mediated Cholesterol Transfer and Ebola Infection
Cell, 165, 2016
|
|
5JDS
 
 | |
5JDR
 
 | | Structure of PD-L1 | | Descriptor: | Programmed cell death 1 ligand 1 | | Authors: | Zhou, A, Wei, H. | | Deposit date: | 2016-04-17 | | Release date: | 2017-04-12 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural basis of a novel PD-L1 nanobody for immune checkpoint blockade.
Cell Discov, 3, 2017
|
|
3TOE
 
 | | Structure of Mth10b | | Descriptor: | DNA/RNA-binding protein Alba | | Authors: | Pan, X.M, Zhang, N, Liu, Y.F, Liu, X. | | Deposit date: | 2011-09-05 | | Release date: | 2012-04-25 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.197 Å) | | Cite: | Molecular mechanism underlying the interaction of typical Sac10b family proteins with DNA.
Plos One, 7, 2012
|
|
7YR0
 
 | | SARS-CoV-2 BA.2.75 S Trimer in complex with S309 (interface) | | Descriptor: | Heavy chain of S309, IGK@ protein, Spike protein S1, ... | | Authors: | Wang, L. | | Deposit date: | 2022-08-08 | | Release date: | 2022-10-19 | | Last modified: | 2022-11-23 | | Method: | ELECTRON MICROSCOPY (3.98 Å) | | Cite: | Characterization of the enhanced infectivity and antibody evasion of Omicron BA.2.75.
Cell Host Microbe, 30, 2022
|
|
7YQU
 
 | | SARS-CoV-2 BA.2.75 S Trimer (3 RBD Down) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein | | Authors: | Wang, L. | | Deposit date: | 2022-08-08 | | Release date: | 2022-10-19 | | Last modified: | 2022-11-23 | | Method: | ELECTRON MICROSCOPY (3.19 Å) | | Cite: | Characterization of the enhanced infectivity and antibody evasion of Omicron BA.2.75.
Cell Host Microbe, 30, 2022
|
|
7YQT
 
 | | SARS-CoV-2 BA.2.75 S Trimer (1 RBD Up) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein | | Authors: | Wang, L. | | Deposit date: | 2022-08-08 | | Release date: | 2022-10-19 | | Last modified: | 2022-11-23 | | Method: | ELECTRON MICROSCOPY (3.45 Å) | | Cite: | Characterization of the enhanced infectivity and antibody evasion of Omicron BA.2.75.
Cell Host Microbe, 30, 2022
|
|
7YQV
 
 | | pH 5.5 SARS-CoV-2 BA.2.75 S Trimer (1 RBD Up) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein | | Authors: | Wang, L. | | Deposit date: | 2022-08-08 | | Release date: | 2022-10-19 | | Last modified: | 2022-11-23 | | Method: | ELECTRON MICROSCOPY (3.58 Å) | | Cite: | Characterization of the enhanced infectivity and antibody evasion of Omicron BA.2.75.
Cell Host Microbe, 30, 2022
|
|
7YR1
 
 | | SARS-CoV-2 BA.2.75 S Trimer in complex with XG2v024 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein, ... | | Authors: | Wang, L. | | Deposit date: | 2022-08-08 | | Release date: | 2022-10-19 | | Last modified: | 2022-11-23 | | Method: | ELECTRON MICROSCOPY (3.62 Å) | | Cite: | Characterization of the enhanced infectivity and antibody evasion of Omicron BA.2.75.
Cell Host Microbe, 30, 2022
|
|
7YR2
 
 | | SARS-CoV-2 BA.2.75 S Trimer in complex with ACE2(state1) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Angiotensin-converting enzyme 2, ... | | Authors: | Wang, L. | | Deposit date: | 2022-08-08 | | Release date: | 2022-10-19 | | Last modified: | 2022-11-23 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Characterization of the enhanced infectivity and antibody evasion of Omicron BA.2.75.
Cell Host Microbe, 30, 2022
|
|
7YQX
 
 | | SARS-CoV-2 BA.2.75 S Trimer in complex with S309 (state1) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, S309 heavy chain, ... | | Authors: | Wang, L. | | Deposit date: | 2022-08-08 | | Release date: | 2022-10-19 | | Last modified: | 2022-11-23 | | Method: | ELECTRON MICROSCOPY (3.72 Å) | | Cite: | Characterization of the enhanced infectivity and antibody evasion of Omicron BA.2.75.
Cell Host Microbe, 30, 2022
|
|
7YR3
 
 | | SARS-CoV-2 BA.2.75 S Trimer in complex with ACE2(state2) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Angiotensin-converting enzyme 2, ... | | Authors: | Wang, L. | | Deposit date: | 2022-08-08 | | Release date: | 2022-10-19 | | Last modified: | 2022-11-23 | | Method: | ELECTRON MICROSCOPY (3.52 Å) | | Cite: | Characterization of the enhanced infectivity and antibody evasion of Omicron BA.2.75.
Cell Host Microbe, 30, 2022
|
|
7YQW
 
 | | SARS-CoV-2 BA.2.75 S Trimer (3 RBD Down) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein, ... | | Authors: | Wang, L. | | Deposit date: | 2022-08-08 | | Release date: | 2022-10-19 | | Last modified: | 2022-11-23 | | Method: | ELECTRON MICROSCOPY (3.51 Å) | | Cite: | Characterization of the enhanced infectivity and antibody evasion of Omicron BA.2.75.
Cell Host Microbe, 30, 2022
|
|
7YQY
 
 | | SARS-CoV-2 BA.2.75 S Trimer in complex with S309 (state2) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, S309 heavy chain, ... | | Authors: | Wang, L. | | Deposit date: | 2022-08-08 | | Release date: | 2022-10-19 | | Last modified: | 2022-11-23 | | Method: | ELECTRON MICROSCOPY (3.74 Å) | | Cite: | Characterization of the enhanced infectivity and antibody evasion of Omicron BA.2.75.
Cell Host Microbe, 30, 2022
|
|
7YQZ
 
 | | SARS-CoV-2 BA.2.75 S Trimer in complex with S309 (state3) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, S309 heavy chain, S309 light chain, ... | | Authors: | Wang, L. | | Deposit date: | 2022-08-08 | | Release date: | 2022-10-19 | | Last modified: | 2022-11-23 | | Method: | ELECTRON MICROSCOPY (3.84 Å) | | Cite: | Characterization of the enhanced infectivity and antibody evasion of Omicron BA.2.75.
Cell Host Microbe, 30, 2022
|
|
7YVL
 
 | | Omicron BA.4/5 SARS-CoV-2 S RBD in complex with TH272 Fab | | Descriptor: | Spike glycoprotein, TH272 Fab heavy chain, TH272 Fab light chain | | Authors: | Guo, Y, Zhang, G, Liang, J, Liu, F, Rao, Z. | | Deposit date: | 2022-08-19 | | Release date: | 2023-06-28 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Discovery and characterization of potent pan-variant SARS-CoV-2 neutralizing antibodies from individuals with Omicron breakthrough infection.
Nat Commun, 14, 2023
|
|
7YVI
 
 | | Omicron BA.4/5 SARS-CoV-2 S in complex with TH236 Fab | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein, TH236 Fab heavy chain, ... | | Authors: | Guo, Y, Zhang, G, Liang, J, Liu, F, Rao, Z. | | Deposit date: | 2022-08-19 | | Release date: | 2023-06-28 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Discovery and characterization of potent pan-variant SARS-CoV-2 neutralizing antibodies from individuals with Omicron breakthrough infection.
Nat Commun, 14, 2023
|
|
