3ANW
 
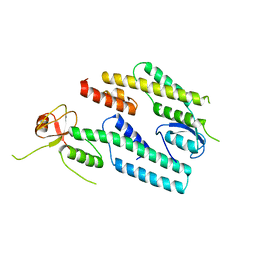 | | A protein complex essential initiation of DNA replication | | Descriptor: | Putative uncharacterized protein | | Authors: | Oyama, T, Shirai, T, Nagasawa, N, Ishino, Y, Morikawa, K. | | Deposit date: | 2010-09-10 | | Release date: | 2011-07-27 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Architectures of archaeal GINS complexes, essential DNA replication initiation factors
Bmc Biol., 9, 2011
|
|
2ZJA
 
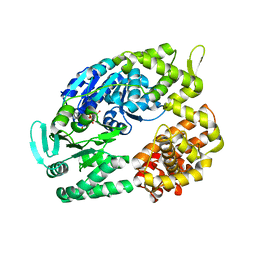 | | Archaeal DNA helicase Hjm complexed with AMPPCP in form 2 | | Descriptor: | PHOSPHOMETHYLPHOSPHONIC ACID ADENYLATE ESTER, Putative ski2-type helicase | | Authors: | Oyama, T, Oka, H, Fujikane, R, Ishino, Y, Morikawa, K. | | Deposit date: | 2008-02-29 | | Release date: | 2009-02-10 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Atomic structures and functional implications of the archaeal RecQ-like helicase Hjm
Bmc Struct.Biol., 9, 2009
|
|
2ZJ2
 
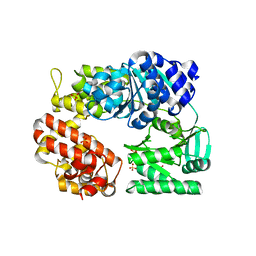 | | Archaeal DNA helicase Hjm apo state in form 1 | | Descriptor: | Putative ski2-type helicase, SULFATE ION | | Authors: | Oyama, T, Oka, H, Fujikane, R, Ishino, Y, Morikawa, K. | | Deposit date: | 2008-02-29 | | Release date: | 2009-02-10 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Atomic structures and functional implications of the archaeal RecQ-like helicase Hjm
Bmc Struct.Biol., 9, 2009
|
|
2ZJ8
 
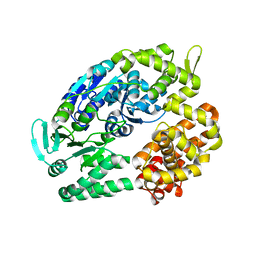 | | Archaeal DNA helicase Hjm apo state in form 2 | | Descriptor: | Putative ski2-type helicase | | Authors: | Oyama, T, Oka, H, Fujikane, R, Ishino, Y, Morikawa, K. | | Deposit date: | 2008-02-29 | | Release date: | 2009-02-10 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Atomic structures and functional implications of the archaeal RecQ-like helicase Hjm
Bmc Struct.Biol., 9, 2009
|
|
2ZJ5
 
 | | Archaeal DNA helicase Hjm complexed with ADP in form 1 | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Putative ski2-type helicase, SULFATE ION | | Authors: | Oyama, T, Oka, H, Fujikane, R, Ishino, Y, Morikawa, K. | | Deposit date: | 2008-02-29 | | Release date: | 2009-02-10 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Atomic structures and functional implications of the archaeal RecQ-like helicase Hjm
Bmc Struct.Biol., 9, 2009
|
|
7E15
 
 | |
4YT1
 
 | | Human PPAR Gamma Ligand Binding Domain in complex with a Gammma Selective Synthetic Partial Agonist MEKT76 | | Descriptor: | N-(benzylsulfonyl)-4-propoxy-3-({[4-(pyrimidin-2-yl)benzoyl]amino}methyl)benzamide, Peroxisome proliferator-activated receptor gamma | | Authors: | Oyama, T, Ohashi, M, Miyachi, H, Kusunoki, M. | | Deposit date: | 2015-03-17 | | Release date: | 2016-03-02 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Peroxisome proliferator-activated receptor gamma (PPAR gamma ) has multiple binding points that accommodate ligands in various conformations: Structurally similar PPAR gamma partial agonists bind to PPAR gamma LBD in different conformations
Bioorg.Med.Chem.Lett., 25, 2015
|
|
3VSO
 
 | | Human PPAR gamma ligand binding domain in complex with a gamma selective agonist mekt21 | | Descriptor: | (2R)-2-benzyl-3-[4-propoxy-3-({[4-(pyrimidin-2-yl)benzoyl]amino}methyl)phenyl]propanoic acid, Peroxisome proliferator-activated receptor gamma | | Authors: | Oyama, T, Waku, T, Ohashi, M, Morikawa, K, Miyachi, H. | | Deposit date: | 2012-04-30 | | Release date: | 2013-05-01 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Design and synthesis of a series of alpha-benzyl phenylpropanoic acid-type peroxisome proliferator-activated receptor (PPAR) gamma partial agonists with improved aqueous solubility
Bioorg.Med.Chem., 21, 2013
|
|
3VI8
 
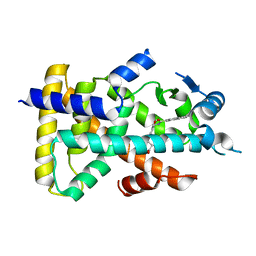 | | Human PPAR alpha ligand binding domain in complex with a synthetic agonist APHM13 | | Descriptor: | (2S)-2-(4-methoxy-3-{[(pyren-1-ylcarbonyl)amino]methyl}benzyl)butanoic acid, Peroxisome proliferator-activated receptor alpha | | Authors: | Oyama, T, Miyachi, H, Morikawa, K. | | Deposit date: | 2011-09-25 | | Release date: | 2012-08-29 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Peroxisome proliferator-activated receptors (PPARs) have multiple binding points that accommodate ligands in various conformations: phenylpropanoic acid-type PPAR ligands bind to PPAR in different conformations, depending on the subtype
J.Med.Chem., 55, 2012
|
|
3AN3
 
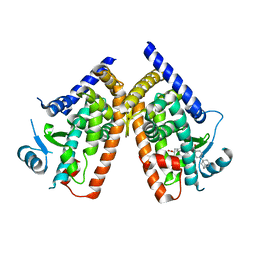 | | Human PPAR gamma ligand binding domain in complex with a gamma selective agonist MO3S | | Descriptor: | (2S)-2-benzyl-3-(4-propoxy-3-{[({4-[(3S,5S,7S)-tricyclo[3.3.1.1~3,7~]dec-1-yl]phenyl}carbonyl)amino]methyl}phenyl)propanoic acid, Peroxisome proliferator-activated receptor gamma | | Authors: | Oyama, T, Ohashi, M, Waku, T, Miyachi, H, Morikawa, K. | | Deposit date: | 2010-08-30 | | Release date: | 2011-07-27 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Design, Synthesis, and Structural Analysis of Phenylpropanoic Acid-Type PPAR gamma-Selective Agonists: Discovery of Reversed Stereochemistry-Activity Relationship
J.Med.Chem., 54, 2011
|
|
3AN4
 
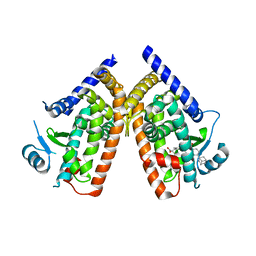 | | Human PPAR gamma ligand binding domain in complex with a gamma selective agonist MO4R | | Descriptor: | (2R)-2-benzyl-3-(4-propoxy-3-{[({4-[(3S,5S,7S)-tricyclo[3.3.1.1~3,7~]dec-1-yl]phenyl}carbonyl)amino]methyl}phenyl)propanoic acid, Peroxisome proliferator-activated receptor gamma | | Authors: | Oyama, T, Ohashi, M, Waku, T, Miyachi, H, Morikawa, K. | | Deposit date: | 2010-08-30 | | Release date: | 2011-07-27 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Design, Synthesis, and Structural Analysis of Phenylpropanoic Acid-Type PPAR gamma-Selective Agonists: Discovery of Reversed Stereochemistry-Activity Relationship
J.Med.Chem., 54, 2011
|
|
6KYP
 
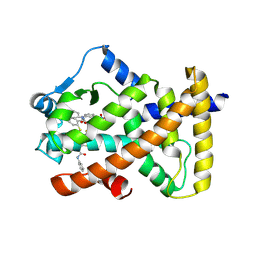 | | X-ray structure of human PPARalpha ligand binding domain-GW9662-clofibric acid co-crystals obtained by delipidation and co-crystallization | | Descriptor: | 2-(4-chloranylphenoxy)-2-methyl-propanoic acid, 2-chloro-5-nitro-N-phenylbenzamide, Peroxisome proliferator-activated receptor alpha | | Authors: | Kamata, S, Saito, K, Honda, A, Ishikawa, R, Oyama, T, Ishii, I. | | Deposit date: | 2019-09-19 | | Release date: | 2020-11-11 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.86 Å) | | Cite: | PPAR alpha Ligand-Binding Domain Structures with Endogenous Fatty Acids and Fibrates.
Iscience, 23, 2020
|
|
6L37
 
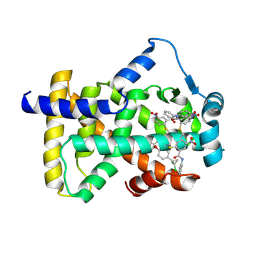 | | X-ray structure of human PPARalpha ligand binding domain-GW9662-ciprofibrate co-crystals obtained by delipidation and co-crystallization | | Descriptor: | 2-chloro-5-nitro-N-phenylbenzamide, 2-{4-[(1S)-2,2-dichlorocyclopropyl]phenoxy}-2-methylpropanoic acid, Peroxisome proliferator-activated receptor alpha | | Authors: | Kamata, S, Saito, K, Honda, A, Ishikawa, R, Oyama, T, Ishii, I. | | Deposit date: | 2019-10-09 | | Release date: | 2020-11-11 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.91 Å) | | Cite: | PPAR alpha Ligand-Binding Domain Structures with Endogenous Fatty Acids and Fibrates.
Iscience, 23, 2020
|
|
6L38
 
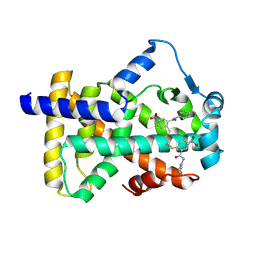 | | X-ray structure of human PPARalpha ligand binding domain-GW9662-gemfibrozil co-crystals obtained by delipidation and co-crystallization | | Descriptor: | 2-chloro-5-nitro-N-phenylbenzamide, Peroxisome proliferator-activated receptor alpha | | Authors: | Kamata, S, Saito, K, Honda, A, Ishikawa, R, Oyama, T, Ishii, I. | | Deposit date: | 2019-10-09 | | Release date: | 2020-11-11 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.761 Å) | | Cite: | PPAR alpha Ligand-Binding Domain Structures with Endogenous Fatty Acids and Fibrates.
Iscience, 23, 2020
|
|
6L36
 
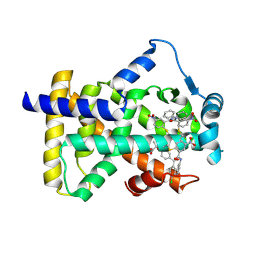 | | X-ray structure of human PPARalpha ligand binding domain-GW9662-fenofibric acid co-crystals obtained by delipidation and co-crystallization | | Descriptor: | 2-[4-(4-chlorobenzene-1-carbonyl)phenoxy]-2-methylpropanoic acid, 2-chloro-5-nitro-N-phenylbenzamide, Peroxisome proliferator-activated receptor alpha | | Authors: | Kamata, S, Saito, K, Honda, A, Ishikawa, R, Oyama, T, Ishii, I. | | Deposit date: | 2019-10-09 | | Release date: | 2020-11-11 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3.301 Å) | | Cite: | PPAR alpha Ligand-Binding Domain Structures with Endogenous Fatty Acids and Fibrates.
Iscience, 23, 2020
|
|
6LX4
 
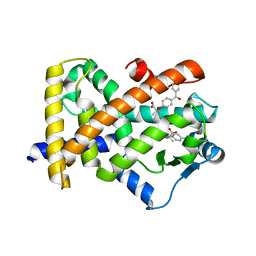 | | X-ray structure of human PPARalpha ligand binding domain-fenofibric acid co-crystals obtained by delipidation and co-crystallization | | Descriptor: | 2-[4-(4-chlorobenzene-1-carbonyl)phenoxy]-2-methylpropanoic acid, Peroxisome proliferator-activated receptor alpha | | Authors: | Kamata, S, Saito, K, Honda, A, Ishikawa, R, Oyama, T, Ishii, I. | | Deposit date: | 2020-02-10 | | Release date: | 2020-11-11 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.13 Å) | | Cite: | PPAR alpha Ligand-Binding Domain Structures with Endogenous Fatty Acids and Fibrates.
Iscience, 23, 2020
|
|
6LX9
 
 | | X-ray structure of human PPARalpha ligand binding domain-arachidonic acid co-crystals obtained by delipidation and cross-seeding | | Descriptor: | ARACHIDONIC ACID, GLYCEROL, Peroxisome proliferator-activated receptor alpha | | Authors: | Kamata, S, Saito, K, Honda, A, Ishikawa, R, Oyama, T, Ishii, I. | | Deposit date: | 2020-02-10 | | Release date: | 2020-11-11 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | PPAR alpha Ligand-Binding Domain Structures with Endogenous Fatty Acids and Fibrates.
Iscience, 23, 2020
|
|
1WDL
 
 | | fatty acid beta-oxidation multienzyme complex from Pseudomonas fragi, form II (native4) | | Descriptor: | 3,6,9,12,15-PENTAOXATRICOSAN-1-OL, 3-ketoacyl-CoA thiolase, ACETYL COENZYME *A, ... | | Authors: | Ishikawa, M, Tsuchiya, D, Oyama, T, Tsunaka, Y, Morikawa, K. | | Deposit date: | 2004-05-17 | | Release date: | 2004-07-27 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Structural basis for channelling mechanism of a fatty acid beta-oxidation multienzyme complex
Embo J., 23, 2004
|
|
1WDK
 
 | | fatty acid beta-oxidation multienzyme complex from Pseudomonas fragi, form I (native2) | | Descriptor: | 3,6,9,12,15-PENTAOXATRICOSAN-1-OL, 3-ketoacyl-CoA thiolase, ACETYL COENZYME *A, ... | | Authors: | Ishikawa, M, Tsuchiya, D, Oyama, T, Tsunaka, Y, Morikawa, K. | | Deposit date: | 2004-05-17 | | Release date: | 2004-07-27 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural basis for channelling mechanism of a fatty acid beta-oxidation multienzyme complex
Embo J., 23, 2004
|
|
1WDM
 
 | | fatty acid beta-oxidation multienzyme complex from Pseudomonas fragi, form I (native3) | | Descriptor: | 3-ketoacyl-CoA thiolase, ACETYL COENZYME *A, Fatty oxidation complex alpha subunit, ... | | Authors: | Ishikawa, M, Tsuchiya, D, Oyama, T, Tsunaka, Y, Morikawa, K. | | Deposit date: | 2004-05-17 | | Release date: | 2004-07-27 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (3.8 Å) | | Cite: | Structural basis for channelling mechanism of a fatty acid beta-oxidation multienzyme complex
Embo J., 23, 2004
|
|
1WWJ
 
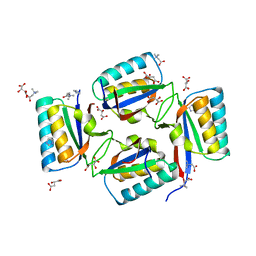 | | crystal structure of KaiB from Synechocystis sp. | | Descriptor: | Circadian clock protein kaiB, D-MALATE, IMIDAZOLE, ... | | Authors: | Hitomi, K, Oyama, T, Han, S, Arvai, A.S, Tainer, J.A, Getzoff, E.D. | | Deposit date: | 2005-01-06 | | Release date: | 2005-02-15 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Tetrameric architecture of the circadian clock protein KaiB. A novel interface for intermolecular interactions and its impact on the circadian rhythm.
J.Biol.Chem., 280, 2005
|
|
1YXJ
 
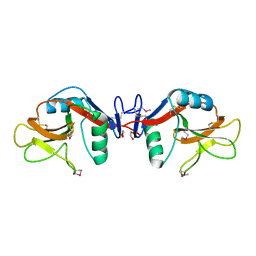 | | Crystal structure of human lectin-like oxidized low-density lipoprotein receptor 1 (LOX-1) at low pH | | Descriptor: | 1,2-ETHANEDIOL, oxidised low density lipoprotein (lectin-like) receptor 1 | | Authors: | Ohki, I, Ishigaki, T, Oyama, T, Matsunaga, S, Xie, Q, Ohnishi-Kameyama, M, Murata, T, Tsuchiya, D, Machida, S, Morikawa, K, Tate, S. | | Deposit date: | 2005-02-22 | | Release date: | 2005-06-14 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | Crystal structure of human lectin-like, oxidized low-density lipoprotein receptor 1 ligand binding domain and its ligand recognition mode to OxLDL.
Structure, 13, 2005
|
|
1YXK
 
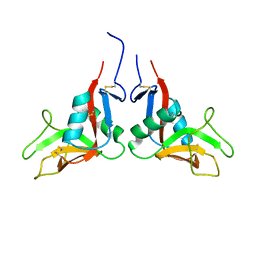 | | Crystal structure of human lectin-like oxidized low-density lipoprotein receptor 1 (LOX-1) disulfide-linked dimer | | Descriptor: | oxidised low density lipoprotein (lectin-like) receptor 1 | | Authors: | Ohki, I, Ishigaki, T, Oyama, T, Matsunaga, S, Xie, Q, Ohnishi-Kameyama, M, Murata, T, Tsuchiya, D, Machida, S, Morikawa, K, Tate, S. | | Deposit date: | 2005-02-22 | | Release date: | 2005-06-14 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structure of human lectin-like, oxidized low-density lipoprotein receptor 1 ligand binding domain and its ligand recognition mode to OxLDL.
Structure, 13, 2005
|
|
7BQ3
 
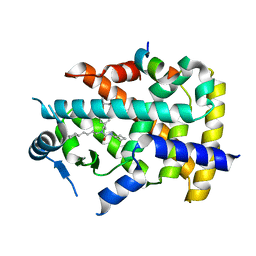 | | X-ray structure of human PPARalpha ligand binding domain-GW7647-SRC1 coactivator peptide co-crystals obtained by delipidation and co-crystallization | | Descriptor: | 15-meric peptide from Nuclear receptor coactivator 1, 2-[(4-{2-[(4-cyclohexylbutyl)(cyclohexylcarbamoyl)amino]ethyl}phenyl)sulfanyl]-2-methylpropanoic acid, Peroxisome proliferator-activated receptor alpha | | Authors: | Kamata, S, Ishikawa, R, Akahane, M, Oyama, T, Ishii, I. | | Deposit date: | 2020-03-23 | | Release date: | 2020-11-11 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | PPAR alpha Ligand-Binding Domain Structures with Endogenous Fatty Acids and Fibrates.
Iscience, 23, 2020
|
|
7BQ4
 
 | | X-ray structure of human PPARalpha ligand binding domain-eicosapentaenoic acid (EPA)-SRC1 coactivator peptide co-crystals obtained by delipidation and co-crystallization | | Descriptor: | 15-meric peptide from Nuclear receptor coactivator 1, 5,8,11,14,17-EICOSAPENTAENOIC ACID, GLYCEROL, ... | | Authors: | Kamata, S, Ishikawa, R, Akahane, M, Oyama, T, Ishii, I. | | Deposit date: | 2020-03-23 | | Release date: | 2020-11-11 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.62 Å) | | Cite: | PPAR alpha Ligand-Binding Domain Structures with Endogenous Fatty Acids and Fibrates.
Iscience, 23, 2020
|
|
