3KYD
 
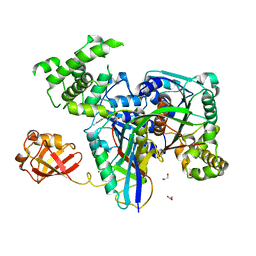 | | Human SUMO E1~SUMO1-AMP tetrahedral intermediate mimic | | Descriptor: | 1,2-ETHANEDIOL, 5'-{[(3-aminopropyl)sulfonyl]amino}-5'-deoxyadenosine, SUMO-activating enzyme subunit 1, ... | | Authors: | Lima, C.D. | | Deposit date: | 2009-12-05 | | Release date: | 2010-02-16 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.61 Å) | | Cite: | Active site remodelling accompanies thioester bond formation in the SUMO E1.
Nature, 463, 2010
|
|
5JNE
 
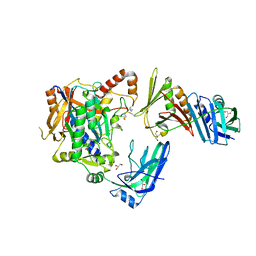 | | E2-SUMO-Siz1 E3-SUMO-PCNA complex | | Descriptor: | E3 SUMO-protein ligase SIZ1,Ubiquitin-like protein SMT3, GLYCEROL, Proliferating cell nuclear antigen, ... | | Authors: | Lima, C.D, Streich Jr, F.C. | | Deposit date: | 2016-04-29 | | Release date: | 2016-08-10 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Capturing a substrate in an activated RING E3/E2-SUMO complex.
Nature, 536, 2016
|
|
5K36
 
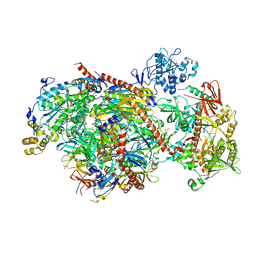 | | Structure of an eleven component nuclear RNA exosome complex bound to RNA | | Descriptor: | Exosome complex component CSL4, Exosome complex component MTR3, Exosome complex component RRP4, ... | | Authors: | Lima, C.D, Zinder, J.C. | | Deposit date: | 2016-05-19 | | Release date: | 2016-11-02 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Nuclear RNA Exosome at 3.1 angstrom Reveals Substrate Specificities, RNA Paths, and Allosteric Inhibition of Rrp44/Dis3.
Mol.Cell, 64, 2016
|
|
6U75
 
 | |
3I2D
 
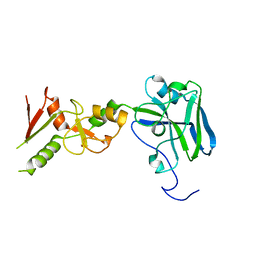 | |
3II4
 
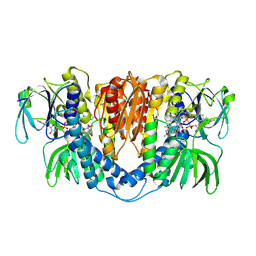 | |
3KYC
 
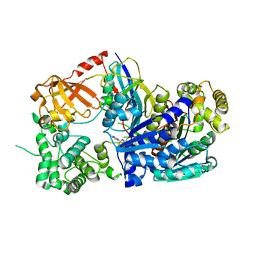 | | Human SUMO E1 complex with a SUMO1-AMP mimic | | Descriptor: | 5'-deoxy-5'-(sulfamoylamino)adenosine, SUMO-activating enzyme subunit 1, SUMO-activating enzyme subunit 2, ... | | Authors: | Lima, C.D. | | Deposit date: | 2009-12-05 | | Release date: | 2010-02-16 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Active site remodelling accompanies thioester bond formation in the SUMO E1.
Nature, 463, 2010
|
|
3KYH
 
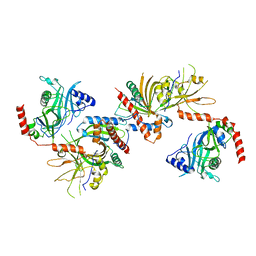 | | Saccharomyces cerevisiae Cet1-Ceg1 capping apparatus | | Descriptor: | mRNA-capping enzyme subunit alpha, mRNA-capping enzyme subunit beta | | Authors: | Lima, C.D. | | Deposit date: | 2009-12-06 | | Release date: | 2010-02-16 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structure of the Saccharomyces cerevisiae Cet1-Ceg1 mRNA Capping Apparatus.
Structure, 18, 2010
|
|
4M52
 
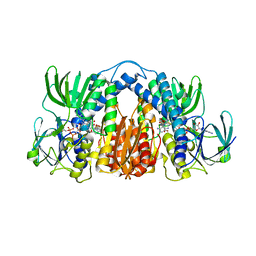 | | Structure of Mtb Lpd bound to SL827 | | Descriptor: | Dihydrolipoyl dehydrogenase, FLAVIN-ADENINE DINUCLEOTIDE, N~2~-[(2-amino-5-bromopyridin-3-yl)sulfonyl]-N-(4-methoxyphenyl)-N~2~-methylglycinamide | | Authors: | Lima, C.D. | | Deposit date: | 2013-08-07 | | Release date: | 2013-11-27 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Lipoamide channel-binding sulfonamides selectively inhibit mycobacterial lipoamide dehydrogenase.
Biochemistry, 52, 2013
|
|
4OO1
 
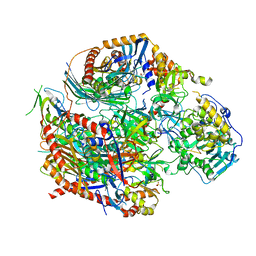 | |
4PN0
 
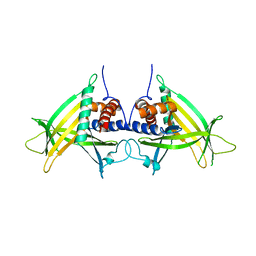 | |
4PN1
 
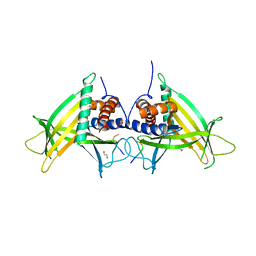 | |
1JR7
 
 | |
2HV9
 
 | | Encephalitozoon cuniculi mRNA Cap (Guanine-N7) Methyltransferase in complex with sinefungin | | Descriptor: | SINEFUNGIN, mRNA cap guanine-N7 methyltransferase | | Authors: | Lima, C.D. | | Deposit date: | 2006-07-27 | | Release date: | 2006-09-19 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Mutational Analysis of Encephalitozoon cuniculi mRNA Cap (Guanine-N7) Methyltransferase, Structure of the Enzyme Bound to Sinefungin, and Evidence That Cap Methyltransferase Is the Target of Sinefungin's Antifungal Activity
J.Biol.Chem., 281, 2006
|
|
5VZJ
 
 | |
7KMY
 
 | | Structure of Mtb Lpd bound to 010705 | | Descriptor: | Dihydrolipoyl dehydrogenase, FLAVIN-ADENINE DINUCLEOTIDE, GLYCEROL, ... | | Authors: | Lima, C.D. | | Deposit date: | 2020-11-03 | | Release date: | 2021-01-27 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.21 Å) | | Cite: | Whole Cell Active Inhibitors of Mycobacterial Lipoamide Dehydrogenase Afford Selectivity over the Human Enzyme through Tight Binding Interactions.
Acs Infect Dis., 7, 2021
|
|
6O82
 
 | | S. pombe ubiquitin E1 complex with a ubiquitin-AMP mimic | | Descriptor: | 1,2-ETHANEDIOL, 5'-deoxy-5'-(sulfamoylamino)adenosine, MAGNESIUM ION, ... | | Authors: | Olsen, S.K, Lima, C.D. | | Deposit date: | 2019-03-08 | | Release date: | 2019-06-19 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.604 Å) | | Cite: | Structural basis for adenylation and thioester bond formation in the ubiquitin E1.
Proc.Natl.Acad.Sci.USA, 116, 2019
|
|
6O83
 
 | | S. pombe ubiquitin E1~ubiquitin-AMP tetrahedral intermediate mimic | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 5'-{[(3-aminopropyl)sulfonyl]amino}-5'-deoxyadenosine, ... | | Authors: | Hann, Z.S, Lima, C.D. | | Deposit date: | 2019-03-08 | | Release date: | 2019-06-19 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (3.153 Å) | | Cite: | Structural basis for adenylation and thioester bond formation in the ubiquitin E1.
Proc.Natl.Acad.Sci.USA, 116, 2019
|
|
6OP8
 
 | | S. pombe Ubc7/U7BR complex | | Descriptor: | 1,2-ETHANEDIOL, CUE domain-containing protein 4, mitochondrial, ... | | Authors: | Hann, Z.S, Lima, C.D. | | Deposit date: | 2019-04-24 | | Release date: | 2019-08-07 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.703 Å) | | Cite: | Crystal structure of the Schizosaccharomyces pombe U7BR E2-binding region in complex with Ubc7.
Acta Crystallogr.,Sect.F, 75, 2019
|
|
4XQ0
 
 | |
1EUV
 
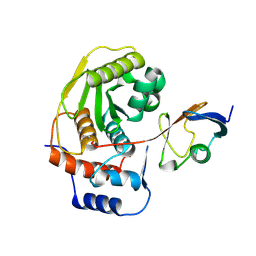 | |
3UIO
 
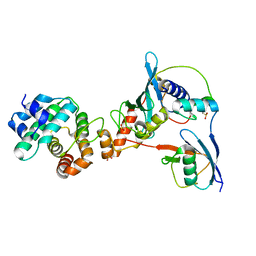 | | Complex between human RanGAP1-SUMO2, UBC9 and the IR1 domain from RanBP2 containing IR2 Motif II | | Descriptor: | E3 SUMO-protein ligase RanBP2, Ran GTPase-activating protein 1, SUMO-conjugating enzyme UBC9, ... | | Authors: | Gareau, J.R, Reverter, D, Lima, C.D. | | Deposit date: | 2011-11-05 | | Release date: | 2011-12-28 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.602 Å) | | Cite: | Determinants of small ubiquitin-like modifier 1 (SUMO1) protein specificity, E3 ligase, and SUMO-RanGAP1 binding activities of nucleoporin RanBP2.
J.Biol.Chem., 287, 2012
|
|
3UIN
 
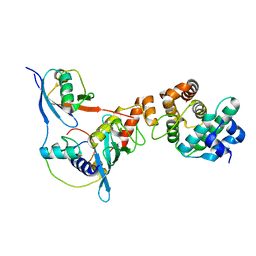 | | Complex between human RanGAP1-SUMO2, UBC9 and the IR1 domain from RanBP2 | | Descriptor: | E3 SUMO-protein ligase RanBP2, Ran GTPase-activating protein 1, SUMO-conjugating enzyme UBC9, ... | | Authors: | Gareau, J.R, Reverter, D, Lima, C.D. | | Deposit date: | 2011-11-05 | | Release date: | 2011-12-28 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.597 Å) | | Cite: | Determinants of small ubiquitin-like modifier 1 (SUMO1) protein specificity, E3 ligase, and SUMO-RanGAP1 binding activities of nucleoporin RanBP2.
J.Biol.Chem., 287, 2012
|
|
3UIP
 
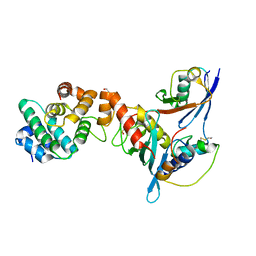 | | Complex between human RanGAP1-SUMO1, UBC9 and the IR1 domain from RanBP2 containing IR2 Motif II | | Descriptor: | E3 SUMO-protein ligase RanBP2, Ran GTPase-activating protein 1, SUMO-conjugating enzyme UBC9, ... | | Authors: | Gareau, J.R, Reverter, D, Lima, C.D. | | Deposit date: | 2011-11-05 | | Release date: | 2011-12-28 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.293 Å) | | Cite: | Determinants of small ubiquitin-like modifier 1 (SUMO1) protein specificity, E3 ligase, and SUMO-RanGAP1 binding activities of nucleoporin RanBP2.
J.Biol.Chem., 287, 2012
|
|
3V62
 
 | | Structure of the S. cerevisiae Srs2 C-terminal domain in complex with PCNA conjugated to SUMO on lysine 164 | | Descriptor: | ATP-dependent DNA helicase SRS2, N-ETHYLMALEIMIDE, Proliferating cell nuclear antigen, ... | | Authors: | Armstrong, A.A, Mohideen, F, Lima, C.D. | | Deposit date: | 2011-12-18 | | Release date: | 2012-02-29 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Recognition of SUMO-modified PCNA requires tandem receptor motifs in Srs2.
Nature, 483, 2012
|
|
