7SG3
 
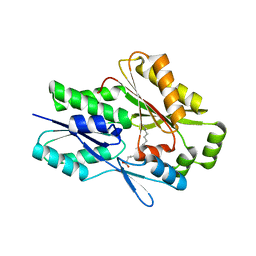 | | The X-ray crystal structure of the Staphylococcus aureus Fatty Acid Kinase B1 mutant A121I-A158L to 2.35 Angstrom resolution (Open form chain A, Palmitate bound) | | Descriptor: | Fatty Acid Kinase B1, PALMITIC ACID | | Authors: | Cuypers, M.G, Gullett, J.M, Rock, C.O, White, S.W. | | Deposit date: | 2021-10-04 | | Release date: | 2021-10-27 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Identification of structural transitions in bacterial fatty acid binding proteins that permit ligand entry and exit at membranes.
J.Biol.Chem., 298, 2022
|
|
7SCL
 
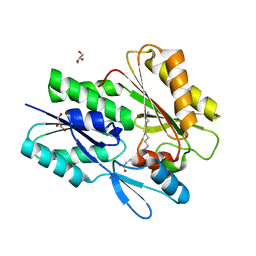 | | The X-ray crystal structure of Staphylococcus aureus Fatty Acid Kinase B1 (FakB1) mutant R173A in complex with Palmitate to 1.60 Angstrom resolution | | Descriptor: | Fatty Acid Kinase B1, GLYCEROL, PALMITIC ACID | | Authors: | Cuypers, M.G, Gullett, J.M, Subramanian, C, Rock, C.O, White, S.W. | | Deposit date: | 2021-09-28 | | Release date: | 2021-10-27 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Identification of structural transitions in bacterial fatty acid binding proteins that permit ligand entry and exit at membranes.
J.Biol.Chem., 298, 2022
|
|
5L9U
 
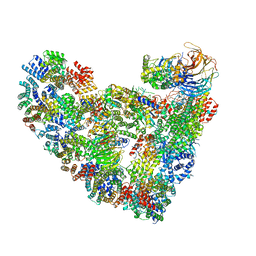 | | Model of human Anaphase-promoting complex/Cyclosome (APC/C-CDH1) with a cross linked Ubiquitin variant-substrate-UBE2C (UBCH10) complex representing key features of multiubiquitination | | Descriptor: | Anaphase-promoting complex subunit 1, Anaphase-promoting complex subunit 10, Anaphase-promoting complex subunit 11, ... | | Authors: | Brown, N.G, VanderLinden, R, Dube, P, Haselbach, D, Peters, J.M, Stark, H, Schulman, B.A. | | Deposit date: | 2016-06-11 | | Release date: | 2016-09-14 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (6.4 Å) | | Cite: | Dual RING E3 Architectures Regulate Multiubiquitination and Ubiquitin Chain Elongation by APC/C.
Cell, 165, 2016
|
|
5L9T
 
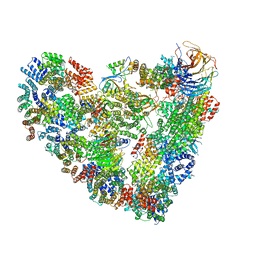 | | Model of human Anaphase-promoting complex/Cyclosome (APC/C-CDH1) with E2 UBE2S poised for polyubiquitination where UBE2S, APC2, and APC11 are modeled into low resolution density | | Descriptor: | Anaphase-promoting complex subunit 1, Anaphase-promoting complex subunit 10, Anaphase-promoting complex subunit 11, ... | | Authors: | Brown, N.G, VanderLinden, R, Dube, P, Haselbach, D, Peters, J.M, Stark, H, Schulman, B.A. | | Deposit date: | 2016-06-11 | | Release date: | 2016-10-26 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (6.4 Å) | | Cite: | Dual RING E3 Architectures Regulate Multiubiquitination and Ubiquitin Chain Elongation by APC/C.
Cell, 165, 2016
|
|
8UR3
 
 | |
8UR6
 
 | |
6MH9
 
 | | The crystal structure of the Staphylococcus aureus Fatty acid Kinase (Fak) B1 protein A121I mutant to 2.02 Angstrom resolution | | Descriptor: | Fatty Acid Kinase (Fak) B1 protein, PALMITIC ACID | | Authors: | Cuypers, M.G, Ericson, M, Subramanian, C, White, S.W, Rock, C.O. | | Deposit date: | 2018-09-17 | | Release date: | 2019-11-13 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.02 Å) | | Cite: | Identification of structural transitions in bacterial fatty acid binding proteins that permit ligand entry and exit at membranes.
J.Biol.Chem., 298, 2022
|
|
6NM1
 
 | | The crystal structure of the Staphylococcus aureus Fatty acid Kinase (Fak) B1 protein A158L mutant to 2.33 Angstrom resolution exhibits a conformation change compared to the wild type form | | Descriptor: | Fatty acid Kinase (Fak) B1 protein, MYRISTIC ACID | | Authors: | Cuypers, M.G, Gullett, J.M, Subramanian, C, Ericson, M, White, S.W, Rock, C.O. | | Deposit date: | 2019-01-10 | | Release date: | 2020-01-15 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.33 Å) | | Cite: | Identification of structural transitions in bacterial fatty acid binding proteins that permit ligand entry and exit at membranes.
J.Biol.Chem., 298, 2022
|
|
7M5C
 
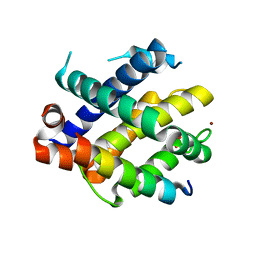 | |
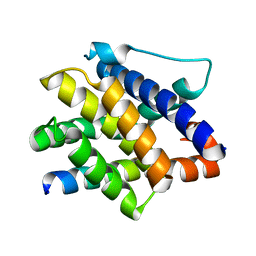
7M5A
 
 | |
7M5B
 
 | |
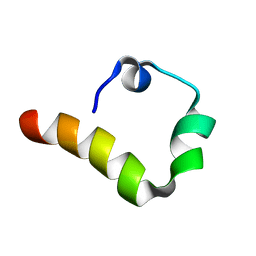
6AXD
 
 | |
6ASR
 
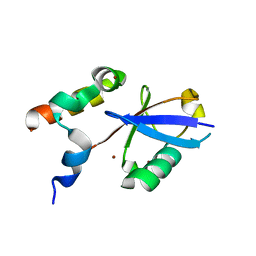 | | REV1 UBM2 domain complex with ubiquitin | | Descriptor: | DNA repair protein REV1, NICKEL (II) ION, Ubiquitin | | Authors: | Miller, D.J. | | Deposit date: | 2017-08-25 | | Release date: | 2018-06-20 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.356 Å) | | Cite: | Structures of REV1 UBM2 Domain Complex with Ubiquitin and with a Small-Molecule that Inhibits the REV1 UBM2-Ubiquitin Interaction.
J. Mol. Biol., 430, 2018
|
|
2M04
 
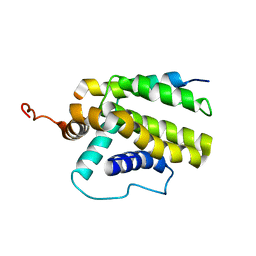 | |
2M03
 
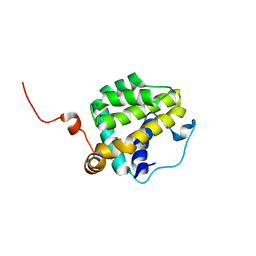 | |
2MT5
 
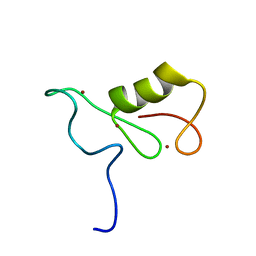 | | Isolated Ring domain | | Descriptor: | Anaphase-promoting complex subunit 11, ZINC ION | | Authors: | Brown, N.G, Watson, E.R, Weissman, F, Royappa, G, Schulman, B, Jarvis, M, Vanderlinden, R, Frye, J.J, Qiao, R, Petzold, G, Peters, J, Stark, H. | | Deposit date: | 2014-08-13 | | Release date: | 2014-10-29 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Mechanism of Polyubiquitination by Human Anaphase-Promoting Complex: RING Repurposing for Ubiquitin Chain Assembly.
Mol.Cell, 56, 2014
|
|
2M6N
 
 | | 3D solution structure of EMI1 (Early Mitotic Inhibitor 1) | | Descriptor: | F-box only protein 5, ZINC ION | | Authors: | Frye, J.J, Brown, N.G, Petzold, G, Watson, E.R, Royappa, G.R, Nourse, A, Jarvis, M, Kriwacki, R.W, Peters, J, Stark, H, Schulman, B.A. | | Deposit date: | 2013-04-06 | | Release date: | 2013-05-29 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Electron microscopy structure of human APC/C(CDH1)-EMI1 reveals multimodal mechanism of E3 ligase shutdown.
Nat.Struct.Mol.Biol., 20, 2013
|
|
2MAH
 
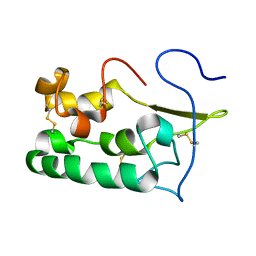 | | Solution structure of Smoothened | | Descriptor: | Protein smoothened | | Authors: | Rana, R, Lee, H, Zheng, J.J. | | Deposit date: | 2013-07-09 | | Release date: | 2014-03-05 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Structural insights into the role of the Smoothened cysteine-rich domain in Hedgehog signalling.
Nat Commun, 4, 2013
|
|
2ME9
 
 | |
2ME8
 
 | |
2MEJ
 
 | |
