5V5G
 
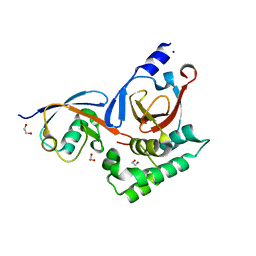 | |
5V5I
 
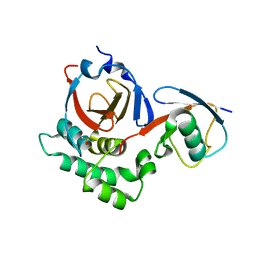 | |
8I71
 
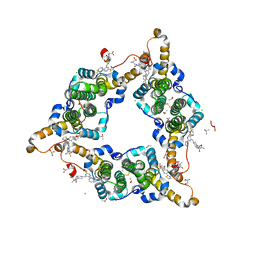 | | Hepatitis B virus core protein Y132A mutant in complex with Linvencorvir (RG7907), a Hepatitis B Virus (HBV) Core Protein Allosteric Modulator (CpAM) | | Descriptor: | 3-[(8~{a}~{S})-7-[[5-ethoxycarbonyl-4-(3-fluoranyl-2-methyl-phenyl)-2-(1,3-thiazol-2-yl)-1,4-dihydropyrimidin-6-yl]methyl]-3-oxidanylidene-5,6,8,8~{a}-tetrahydro-1~{H}-imidazo[1,5-a]pyrazin-2-yl]-2,2-dimethyl-propanoic acid, CHLORIDE ION, Capsid protein, ... | | Authors: | Zhou, Z, Xu, Z.H. | | Deposit date: | 2023-01-30 | | Release date: | 2023-03-22 | | Last modified: | 2023-04-05 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Discovery of Linvencorvir (RG7907), a Hepatitis B Virus Core Protein Allosteric Modulator, for the Treatment of Chronic HBV Infection.
J.Med.Chem., 66, 2023
|
|
3IUF
 
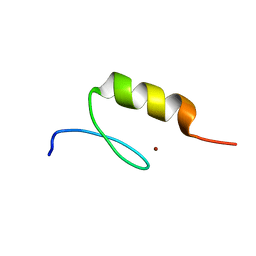 | | Crystal structure of the C2H2-type zinc finger domain of human ubi-d4 | | Descriptor: | ZINC ION, Zinc finger protein ubi-d4 | | Authors: | Tempel, W, Xu, C, Bian, C, Adams-Cioaba, M, Eryilmaz, J, Bountra, C, Weigelt, J, Arrowsmith, C.H, Edwards, A.M, Bochkarev, A, Min, J, Structural Genomics Consortium (SGC) | | Deposit date: | 2009-08-31 | | Release date: | 2009-11-03 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of the Cys2His2-type zinc finger domain of human DPF2.
Biochem.Biophys.Res.Commun., 413, 2011
|
|
8D95
 
 | |
8DEA
 
 | |
8DG6
 
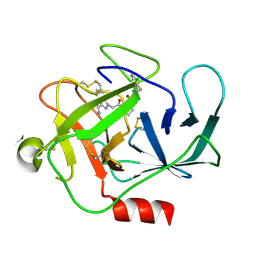 | |
8SRO
 
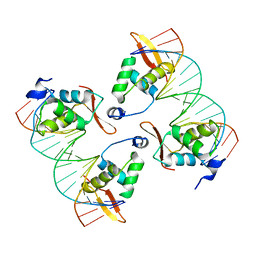 | | FoxP3 tetramer on TTTG repeats | | Descriptor: | DNA 72-mer, Forkhead box protein P3 | | Authors: | Leng, F, Hur, S. | | Deposit date: | 2023-05-05 | | Release date: | 2023-10-18 | | Last modified: | 2023-12-27 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | FOXP3 recognizes microsatellites and bridges DNA through multimerization.
Nature, 624, 2023
|
|
8SRP
 
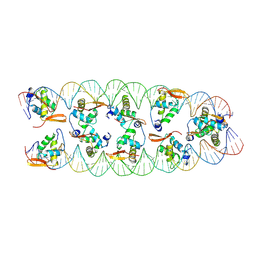 | |
3B6Q
 
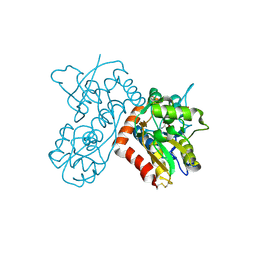 | |
3B6W
 
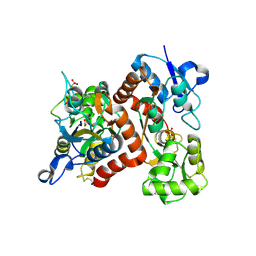 | |
8JNI
 
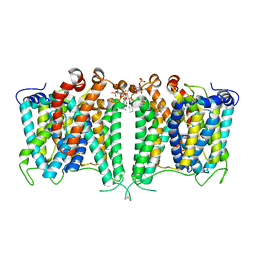 | | Structure of AE2 in complex with PIP2 | | Descriptor: | Anion exchange protein 2, CHLORIDE ION, [(2R)-1-octadecanoyloxy-3-[oxidanyl-[(1R,2R,3S,4R,5R,6S)-2,3,6-tris(oxidanyl)-4,5-diphosphonooxy-cyclohexyl]oxy-phospho ryl]oxy-propan-2-yl] (8Z)-icosa-5,8,11,14-tetraenoate | | Authors: | Yin, Y.X, Ding, D. | | Deposit date: | 2023-06-06 | | Release date: | 2024-02-07 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Structural and functional insights into the lipid regulation of human anion exchanger 2.
Nat Commun, 15, 2024
|
|
8JNJ
 
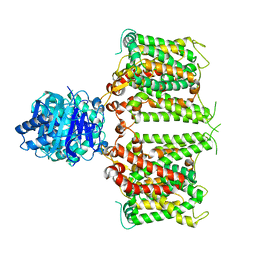 | | Structure of R932A/K1147A/H1148A mutant AE2 | | Descriptor: | Anion exchange protein 2 | | Authors: | Yin, Y.X, Ding, D. | | Deposit date: | 2023-06-06 | | Release date: | 2024-02-07 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Structural and functional insights into the lipid regulation of human anion exchanger 2.
Nat Commun, 15, 2024
|
|
5C7J
 
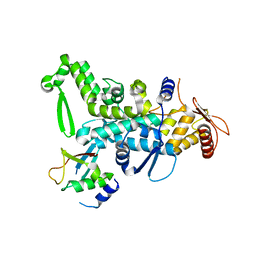 | | CRYSTAL STRUCTURE OF NEDD4 WITH A UB VARIANT | | Descriptor: | E3 ubiquitin-protein ligase NEDD4, Polyubiquitin-C | | Authors: | Walker, J.R, Hu, J, Dong, A, Bountra, C, Edwards, A.M, Arrowsmith, C.H, Tong, Y, Structural Genomics Consortium (SGC) | | Deposit date: | 2015-06-24 | | Release date: | 2016-03-16 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | System-Wide Modulation of HECT E3 Ligases with Selective Ubiquitin Variant Probes.
Mol.Cell, 62, 2016
|
|
5HPS
 
 | |
5HPK
 
 | | System-wide modulation of HECT E3 ligases with selective ubiquitin variant probes: NEDD4L and UbV NL.1 | | Descriptor: | E3 ubiquitin-protein ligase NEDD4-like, Ubiquitin variant NL.1 | | Authors: | Wu, K.-P, Mukherjee, M, Mercredi, P.Y, Schulman, B.A. | | Deposit date: | 2016-01-20 | | Release date: | 2016-03-16 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.431 Å) | | Cite: | System-Wide Modulation of HECT E3 Ligases with Selective Ubiquitin Variant Probes.
Mol.Cell, 62, 2016
|
|
5HPT
 
 | |
5HPL
 
 | |
7EQ2
 
 | | Crystal structure of GDP-bound Rab1a-T75D | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, CACODYLATE ION, COBALT HEXAMMINE(III), ... | | Authors: | Cao, Y.L, Gu, D.D, Gao, S. | | Deposit date: | 2021-04-28 | | Release date: | 2022-11-02 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.55090284 Å) | | Cite: | Aurora kinase A-mediated phosphorylation triggers structural alteration of Rab1A to enhance ER complexity during mitosis
Nat.Struct.Mol.Biol., 2024
|
|
4Y07
 
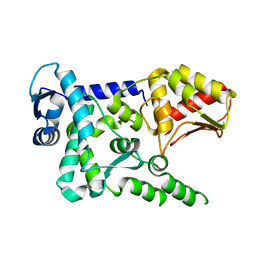 | | Crystal structure of the HECT domain of human WWP2 | | Descriptor: | NEDD4-like E3 ubiquitin-protein ligase WWP2 | | Authors: | Gong, W, Li, J, Li, Z, Xu, Y. | | Deposit date: | 2015-02-05 | | Release date: | 2015-11-04 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.507 Å) | | Cite: | Structure of the HECT domain of human WWP2
Acta Crystallogr.,Sect.F, 71, 2015
|
|
7YR5
 
 | |
4XNW
 
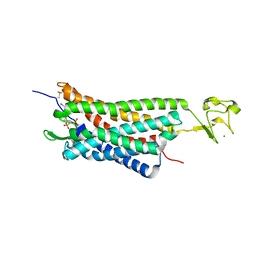 | | The human P2Y1 receptor in complex with MRS2500 | | Descriptor: | P2Y purinoceptor 1,Rubredoxin,P2Y purinoceptor 1, ZINC ION, [(1R,2S,4S,5S)-4-[2-iodo-6-(methylamino)-9H-purin-9-yl]-2-(phosphonooxy)bicyclo[3.1.0]hex-1-yl]methyl dihydrogen phosphate | | Authors: | Zhang, D, Gao, Z, Jacobson, K, Han, G.W, Stevens, R, Zhao, Q, Wu, B, GPCR Network (GPCR) | | Deposit date: | 2015-01-16 | | Release date: | 2015-04-01 | | Last modified: | 2020-02-05 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Two disparate ligand-binding sites in the human P2Y1 receptor
Nature, 520, 2015
|
|
4WIV
 
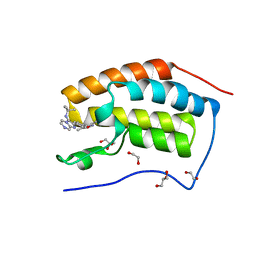 | | Crystal Structure of the first bromodomain of human BRD4 in complex with a novel inhibitor UMB32 (N-TERT-BUTYL-2-[4-(3,5-DIMETHYL-1,2-OXAZOL-4-YL) PHENYL]IMIDAZO[1,2-A]PYRAZIN-3-AMINE) | | Descriptor: | 1,2-ETHANEDIOL, Bromodomain-containing protein 4, GLYCEROL, ... | | Authors: | Xu, X, Blacklow, S. | | Deposit date: | 2014-09-26 | | Release date: | 2014-10-29 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.56 Å) | | Cite: | Biased multicomponent reactions to develop novel bromodomain inhibitors.
J.Med.Chem., 57, 2014
|
|
6PWW
 
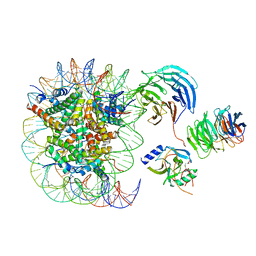 | | Cryo-EM structure of MLL1 in complex with RbBP5 and WDR5 bound to the nucleosome | | Descriptor: | DNA (146-MER), Histone H2A type 1, Histone H2B 1.1, ... | | Authors: | Park, S.H, Ayoub, A, Lee, Y.T, Xu, J, Zhang, W, Zhang, B, Zhang, Y, Cianfrocco, M.A, Su, M, Dou, Y, Cho, U. | | Deposit date: | 2019-07-23 | | Release date: | 2019-12-18 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (4.4 Å) | | Cite: | Cryo-EM structure of the human MLL1 core complex bound to the nucleosome.
Nat Commun, 10, 2019
|
|
6PWX
 
 | | Cryo-EM structure of RbBP5 bound to the nucleosome | | Descriptor: | DNA (146-MER), Histone H2A type 1, Histone H2B 1.1, ... | | Authors: | Park, S.H, Ayoub, A, Lee, Y.T, Xu, J, Zhang, W, Zhang, B, Zhang, Y, Cianfrocco, M.A, Su, M, Dou, Y, Cho, U. | | Deposit date: | 2019-07-23 | | Release date: | 2019-12-18 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (4.2 Å) | | Cite: | Cryo-EM structure of the human MLL1 core complex bound to the nucleosome.
Nat Commun, 10, 2019
|
|
