1TJV
 
 | | Crystal Structure of T161D Duck Delta 2 Crystallin Mutant | | Descriptor: | Delta crystallin II | | Authors: | Sampaleanu, L.M, Codding, P.W, Lobsanov, Y.D, Tsai, M, Smith, G.D, Horvatin, C, Howell, P.L. | | Deposit date: | 2004-06-07 | | Release date: | 2004-09-07 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural studies of duck delta2 crystallin mutants provide insight into the role of Thr161 and the 280s loop in catalysis
BIOCHEM.J., 384, 2004
|
|
5VJU
 
 | | De Novo Photosynthetic Reaction Center Protein Variant Equipped with His-Tyr H-bond, Heme B, and Cd(II) ions | | Descriptor: | CADMIUM ION, PROTOPORPHYRIN IX CONTAINING FE, Reaction Center Maquette Leu71His variant | | Authors: | Ennist, N.M, Stayrook, S.E, Dutton, P.L, Moser, C.C. | | Deposit date: | 2017-04-19 | | Release date: | 2018-04-25 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.08 Å) | | Cite: | De novo protein design of photochemical reaction centers.
Nat Commun, 13, 2022
|
|
1TJW
 
 | | Crystal Structure of T161D Duck Delta 2 Crystallin Mutant with bound argininosuccinate | | Descriptor: | ARGININOSUCCINATE, Delta crystallin II | | Authors: | Sampaleanu, L.M, Codding, P.W, Lobsanov, Y.D, Tsai, M, Smith, G.D, Horvatin, C, Howell, P.L. | | Deposit date: | 2004-06-07 | | Release date: | 2004-09-07 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural studies of duck delta2 crystallin mutants provide insight into the role of Thr161 and the 280s loop in catalysis
Biochem.J., 384, 2004
|
|
5WFT
 
 | | PelB 319-436 from Pseudomonas aeruginosa PAO1 | | Descriptor: | PelB | | Authors: | Marmont, L.S, Howell, P.L. | | Deposit date: | 2017-07-12 | | Release date: | 2017-10-04 | | Last modified: | 2020-01-08 | | Method: | X-RAY DIFFRACTION (2.821 Å) | | Cite: | PelA and PelB proteins form a modification and secretion complex essential for Pel polysaccharide-dependent biofilm formation in Pseudomonas aeruginosa.
J. Biol. Chem., 292, 2017
|
|
1TBW
 
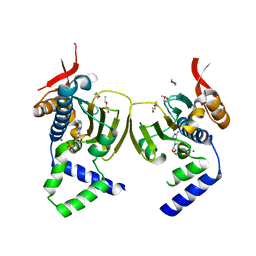 | | Ligand Induced Conformational Shift in the N-terminal Domain of GRP94, Open Conformation | | Descriptor: | ADENOSINE MONOPHOSPHATE, Endoplasmin, MAGNESIUM ION, ... | | Authors: | Gewirth, D.T, Immormino, R.M, Dollins, D.E, Shaffer, P.L, Walker, M.A, Soldano, K.L. | | Deposit date: | 2004-05-20 | | Release date: | 2004-08-24 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Ligand-induced Conformational Shift in the N-terminal Domain of GRP94, an Hsp90 Chaperone.
J.Biol.Chem., 279, 2004
|
|
1TC0
 
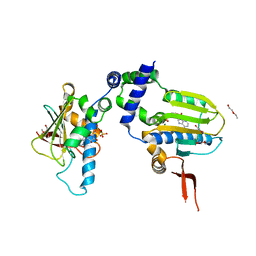 | | Ligand Induced Conformational Shifts in the N-terminal Domain of GRP94, Open Conformation Complexed with the physiological partner ATP | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Endoplasmin, MAGNESIUM ION, ... | | Authors: | Gewirth, D.T, Immormino, R.M, Dollins, D.E, Shaffer, P.L, Walker, M.A, Soldano, K.L. | | Deposit date: | 2004-05-20 | | Release date: | 2004-08-24 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Ligand-induced Conformational Shift in the N-terminal Domain of GRP94, an Hsp90 Chaperone.
J.Biol.Chem., 279, 2004
|
|
1TJU
 
 | | Crystal Structure of T161S Duck Delta 2 Crystallin Mutant | | Descriptor: | Delta crystallin II | | Authors: | Sampaleanu, L.M, Codding, P.W, Lobsanov, Y.D, Tsai, M, Smith, G.D, Horvatin, C, Howell, P.L. | | Deposit date: | 2004-06-07 | | Release date: | 2004-09-07 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural studies of duck delta2 crystallin mutants provide insight into the role of Thr161 and the 280s loop in catalysis
Biochem.J., 384, 2004
|
|
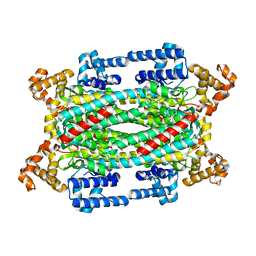
1KP2
 
 | |
1LQ7
 
 | | De Novo Designed Protein Model of Radical Enzymes | | Descriptor: | Alpha3W | | Authors: | Dai, Q.-H, Tommos, C, Fuentes, E.J, Blomberg, M, Dutton, P.L, Wand, A.J. | | Deposit date: | 2002-05-09 | | Release date: | 2002-06-05 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Structure of a De Novo Designed Protein Model of Radical Enzymes
J.Am.Chem.Soc., 124, 2002
|
|
1KP3
 
 | |
6UGM
 
 | | Structural basis of COMPASS eCM recognition of an unmodified nucleosome | | Descriptor: | Bre2, DNA (146-MER), H3 N-terminus, ... | | Authors: | Hsu, P.L, Shi, H, Zheng, N. | | Deposit date: | 2019-09-26 | | Release date: | 2019-11-20 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Structural Basis of H2B Ubiquitination-Dependent H3K4 Methylation by COMPASS.
Mol.Cell, 76, 2019
|
|
6VE4
 
 | | Pentadecameric PilQ from Pseudomonas aeruginosa | | Descriptor: | Fimbrial assembly protein PilQ | | Authors: | McCallum, M, Tammam, S, Rubinstein, J.L, Burrows, L.L, Howell, P.L. | | Deposit date: | 2019-12-28 | | Release date: | 2020-12-23 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (6.9 Å) | | Cite: | CryoEM map of Pseudomonas aeruginosa PilQ enables structural characterization of TsaP.
Structure, 29, 2021
|
|
6VE2
 
 | | Tetradecameric PilQ bound by TsaP heptamer from Pseudomonas aeruginosa | | Descriptor: | Fimbrial assembly protein PilQ, LysM domain-containing protein | | Authors: | McCallum, M, Tammam, S, Rubinstein, J.L, Burrows, L.L, Howell, P.L. | | Deposit date: | 2019-12-28 | | Release date: | 2020-12-23 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (4.3 Å) | | Cite: | CryoEM map of Pseudomonas aeruginosa PilQ enables structural characterization of TsaP.
Structure, 29, 2021
|
|
6VE3
 
 | | Tetradecameric PilQ from Pseudomonas aeruginosa | | Descriptor: | Fimbrial assembly protein PilQ | | Authors: | McCallum, M, Tammam, S, Rubinstein, J.L, Burrows, L.L, Howell, P.L. | | Deposit date: | 2019-12-28 | | Release date: | 2020-12-23 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (4.3 Å) | | Cite: | CryoEM map of Pseudomonas aeruginosa PilQ enables structural characterization of TsaP.
Structure, 29, 2021
|
|
6VJP
 
 | | Structure of Staphylococcus aureus peptidoglycan O-acetyltransferase A (OatA) C-terminal catalytic domain | | Descriptor: | Acetyltransferase, SODIUM ION | | Authors: | Jones, C.J, Sychantha, D, Howell, P.L, Clarke, A.J. | | Deposit date: | 2020-01-16 | | Release date: | 2020-05-06 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.711 Å) | | Cite: | Structural basis for theO-acetyltransferase function of the extracytoplasmic domain of OatA fromStaphylococcus aureus.
J.Biol.Chem., 295, 2020
|
|
6WJA
 
 | |
6WN9
 
 | | Structure of Staphylococcus aureus peptidoglycan O-acetyltransferase A (OatA) C-terminal catalytic domain, Zn-bound | | Descriptor: | Acetyltransferase, ZINC ION | | Authors: | Jones, C.J, Sychantha, D, Howell, P.L, Clarke, A.J. | | Deposit date: | 2020-04-22 | | Release date: | 2020-05-06 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Structural basis for theO-acetyltransferase function of the extracytoplasmic domain of OatA fromStaphylococcus aureus.
J.Biol.Chem., 295, 2020
|
|
6WJ9
 
 | | UDP-GlcNAc C4-epimerase mutant S121A/Y146F from Pseudomonas protegens in complex with UDP-GlcNAc | | Descriptor: | NAD-dependent epimerase/dehydratase family protein, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, URIDINE-DIPHOSPHATE-N-ACETYLGLUCOSAMINE | | Authors: | Marmont, L.S, Willams, R.J, Whitney, J.C, Whitfield, G.B, Robinson, H, Parsek, M.R, Nitz, M, Harrison, J.J, Howell, P.L. | | Deposit date: | 2020-04-13 | | Release date: | 2020-07-08 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.11 Å) | | Cite: | PelX is a UDP-N-acetylglucosamine C4-epimerase involved in Pel polysaccharide-dependent biofilm formation.
J.Biol.Chem., 295, 2020
|
|
6WJB
 
 | | UDP-GlcNAc C4-epimerase from Pseudomonas protegens in complex with NAD and UDP-GlcNAc | | Descriptor: | NAD-dependent epimerase/dehydratase family protein, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, URIDINE-DIPHOSPHATE-N-ACETYLGLUCOSAMINE | | Authors: | Marmont, L.S, Pfoh, R, Robinson, H, Howell, P.L. | | Deposit date: | 2020-04-13 | | Release date: | 2020-07-08 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | PelX is a UDP-N-acetylglucosamine C4-epimerase involved in Pel polysaccharide-dependent biofilm formation.
J.Biol.Chem., 295, 2020
|
|
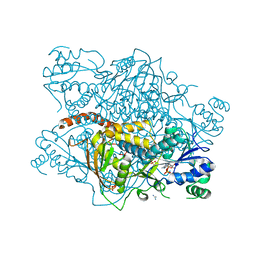
1DF0
 
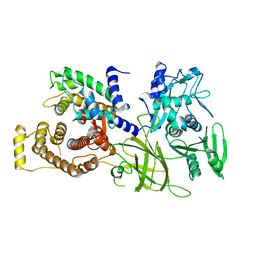 | | Crystal structure of M-Calpain | | Descriptor: | CALPAIN, M-CALPAIN | | Authors: | Hosfield, C.M, Elce, J.S, Davies, P.L, Jia, Z. | | Deposit date: | 1999-11-16 | | Release date: | 2000-06-21 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal structure of calpain reveals the structural basis for Ca(2+)-dependent protease activity and a novel mode of enzyme activation.
EMBO J., 18, 1999
|
|
1EWW
 
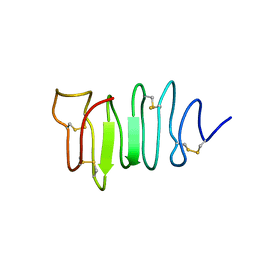 | | SOLUTION STRUCTURE OF SPRUCE BUDWORM ANTIFREEZE PROTEIN AT 30 DEGREES CELSIUS | | Descriptor: | ANTIFREEZE PROTEIN | | Authors: | Graether, S.P, Kuiper, M.J, Gagne, S.M, Walker, V.K, Jia, Z, Sykes, B.D, Davies, P.L. | | Deposit date: | 2000-04-27 | | Release date: | 2000-07-27 | | Last modified: | 2022-02-16 | | Method: | SOLUTION NMR | | Cite: | Beta-helix structure and ice-binding properties of a hyperactive antifreeze protein from an insect.
Nature, 406, 2000
|
|
4NK6
 
 | |
4OZX
 
 | |
4OZZ
 
 | |
4OCW
 
 | | Crystal structure of human Fab CAP256-VRC26.06, a potent V1V2-directed HIV-1 neutralizing antibody | | Descriptor: | CAP256-VRC26.06 heavy chain, CAP256-VRC26.06 light chain | | Authors: | Gorman, J, Doria-Rose, N.A, Schramm, C.A, Moore, P.L, Mascola, J.R, Shapiro, L, Morris, L, Kwong, P.D. | | Deposit date: | 2014-01-09 | | Release date: | 2014-02-26 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (3.001 Å) | | Cite: | Developmental pathway for potent V1V2-directed HIV-neutralizing antibodies.
Nature, 509, 2014
|
|
