1TEW
 
 | |
1EWW
 
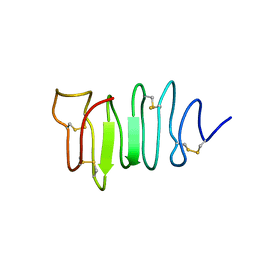 | | SOLUTION STRUCTURE OF SPRUCE BUDWORM ANTIFREEZE PROTEIN AT 30 DEGREES CELSIUS | | Descriptor: | ANTIFREEZE PROTEIN | | Authors: | Graether, S.P, Kuiper, M.J, Gagne, S.M, Walker, V.K, Jia, Z, Sykes, B.D, Davies, P.L. | | Deposit date: | 2000-04-27 | | Release date: | 2000-07-27 | | Last modified: | 2022-02-16 | | Method: | SOLUTION NMR | | Cite: | Beta-helix structure and ice-binding properties of a hyperactive antifreeze protein from an insect.
Nature, 406, 2000
|
|
1UH5
 
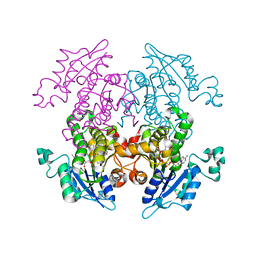 | | Crystal Structure of Enoyl-ACP Reductase with Triclosan at 2.2angstroms | | Descriptor: | NICOTINAMIDE-ADENINE-DINUCLEOTIDE, TRICLOSAN, enoyl-ACP reductase | | Authors: | Swarnamukhi, P.L, Kapoor, M, Surolia, N, Surolia, A, Suguna, K. | | Deposit date: | 2003-06-24 | | Release date: | 2004-09-28 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural basis for the variation in triclosan affinity to enoyl reductases.
J.Mol.Biol., 343, 2004
|
|
1V35
 
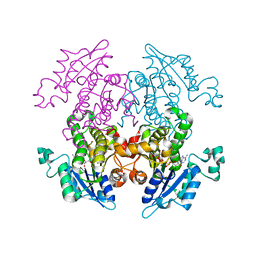 | | Crystal Structure of Eoyl-ACP Reductase with NADH | | Descriptor: | 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, enoyl-ACP reductase | | Authors: | SwarnaMukhi, P.L, Kapoor, M, surolia, N, Surolia, A, Suguna, K. | | Deposit date: | 2003-10-28 | | Release date: | 2004-09-28 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural basis for the variation in triclosan affinity to enoyl reductases.
J.Mol.Biol., 343, 2004
|
|
2OKH
 
 | | Crystal structure of dimeric form of PfFabZ in crystal form3 | | Descriptor: | Beta-hydroxyacyl-ACP dehydratase | | Authors: | Swarnamukhi, P.L, Sharma, S.K, Padala, P, Surolia, N, Surolia, A, Suguna, K. | | Deposit date: | 2007-01-16 | | Release date: | 2007-04-10 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Packing and loop-structure variations in non-isomorphous crystals of FabZ from Plasmodium falciparum
ACTA CRYSTALLOGR.,SECT.D, 63, 2007
|
|
2O8F
 
 | | human MutSalpha (MSH2/MSH6) bound to DNA with a single base T insert | | Descriptor: | 5'-D(*CP*GP*CP*TP*AP*GP*CP*GP*TP*GP*CP*GP*GP*CP*CP*GP*TP*C)-3', 5'-D(*GP*AP*CP*GP*GP*CP*CP*GP*CP*CP*GP*CP*TP*AP*GP*CP*G)-3', ADENOSINE-5'-DIPHOSPHATE, ... | | Authors: | Warren, J.J, Pohlhaus, T.J, Changela, A, Modrich, P.L, Beese, L.S. | | Deposit date: | 2006-12-12 | | Release date: | 2007-06-05 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (3.25 Å) | | Cite: | Structure of the Human MutSalpha DNA Lesion Recognition Complex.
Mol.Cell, 26, 2007
|
|
2O8B
 
 | | human MutSalpha (MSH2/MSH6) bound to ADP and a G T mispair | | Descriptor: | 5'-D(*CP*CP*TP*AP*GP*CP*GP*TP*GP*CP*GP*GP*TP*TP*C)-3', 5'-D(*GP*AP*AP*CP*CP*GP*CP*GP*CP*GP*CP*TP*AP*GP*G)-3', ADENOSINE-5'-DIPHOSPHATE, ... | | Authors: | Warren, J.J, Pohlhaus, T.J, Changela, A, Modrich, P.L, Beese, L.S. | | Deposit date: | 2006-12-12 | | Release date: | 2007-06-05 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Structure of the Human MutSalpha DNA Lesion Recognition Complex.
Mol.Cell, 26, 2007
|
|
2O8E
 
 | | human MutSalpha (MSH2/MSH6) bound to a G T mispair, with ADP bound to MSH2 only | | Descriptor: | 5'-D(*CP*CP*TP*AP*GP*CP*CP*TP*GP*CP*GP*GP*TP*TP*C)-3', 5'-D(*GP*AP*AP*CP*CP*GP*CP*GP*GP*GP*CP*TP*AP*GP*G)-3', ADENOSINE-5'-DIPHOSPHATE, ... | | Authors: | Warren, J.J, Pohlhaus, T.J, Changela, A, Modrich, P.L, Beese, L.S. | | Deposit date: | 2006-12-12 | | Release date: | 2007-06-05 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Structure of the Human MutSalpha DNA Lesion Recognition Complex.
Mol.Cell, 26, 2007
|
|
2O8C
 
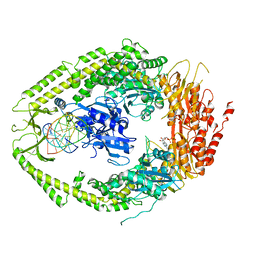 | | human MutSalpha (MSH2/MSH6) bound to ADP and an O6-methyl-guanine T mispair | | Descriptor: | 5'-D(*CP*CP*TP*AP*GP*CP*GP*TP*GP*CP*GP*GP*TP*TP*C)-3', 5'-D(*GP*AP*AP*CP*CP*GP*CP*(6OG)P*CP*GP*CP*TP*AP*GP*G)-3', ADENOSINE-5'-DIPHOSPHATE, ... | | Authors: | Warren, J.J, Pohlhaus, T.J, Changela, A, Modrich, P.L, Beese, L.S. | | Deposit date: | 2006-12-12 | | Release date: | 2007-06-05 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (3.37 Å) | | Cite: | Structure of the Human MutSalpha DNA Lesion Recognition Complex.
Mol.Cell, 26, 2007
|
|
2OKI
 
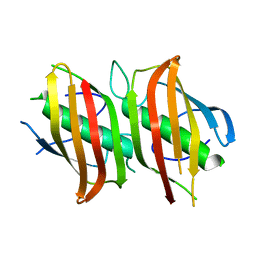 | | Crystal structure of dimeric form of PfFabZ in crystal form2 | | Descriptor: | Beta-hydroxyacyl-ACP dehydratase | | Authors: | Swarnamukhi, P.L, Sharma, S.K, Padala, P, Surolia, N, Surolia, A, Suguna, K. | | Deposit date: | 2007-01-17 | | Release date: | 2007-04-10 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Packing and loop-structure variations in non-isomorphous crystals of FabZ from Plasmodium falciparum
ACTA CRYSTALLOGR.,SECT.D, 63, 2007
|
|
2O8D
 
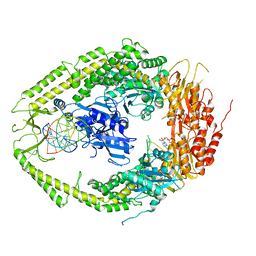 | | human MutSalpha (MSH2/MSH6) bound to ADP and a G dU mispair | | Descriptor: | 5'-D(*CP*CP*TP*AP*GP*CP*GP*(DU)P*GP*CP*GP*GP*TP*TP*C)-3', 5'-D(*GP*AP*AP*CP*CP*GP*CP*GP*CP*GP*CP*TP*AP*GP*G)-3', ADENOSINE-5'-DIPHOSPHATE, ... | | Authors: | Warren, J.J, Pohlhaus, T.J, Changela, A, Modrich, P.L, Beese, L.S. | | Deposit date: | 2006-12-12 | | Release date: | 2007-06-05 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structure of the Human MutSalpha DNA Lesion Recognition Complex.
Mol.Cell, 26, 2007
|
|
2OLC
 
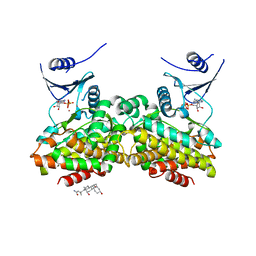 | | Crystal structure of 5-methylthioribose kinase in complex with ADP-2Ho | | Descriptor: | 3-[(3-CHOLAMIDOPROPYL)DIMETHYLAMMONIO]-1-PROPANESULFONATE, ADENOSINE-5'-DIPHOSPHATE, HOLMIUM ATOM, ... | | Authors: | Ku, S.Y, Smith, G.D, Howell, P.L. | | Deposit date: | 2007-01-18 | | Release date: | 2007-05-22 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | ADP-2Ho as a phasing tool for nucleotide-containing proteins.
Acta Crystallogr.,Sect.D, 63, 2007
|
|
2PTQ
 
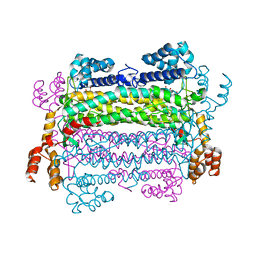 | |
2PTR
 
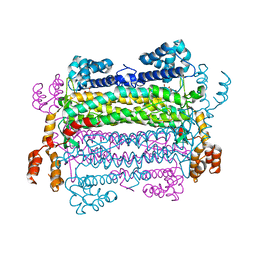 | |
2PTS
 
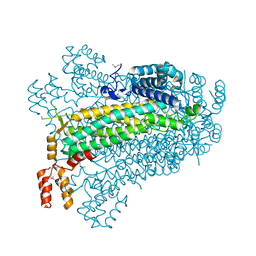 | |
2QSU
 
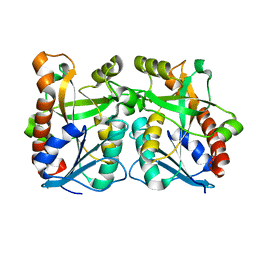 | |
2QTG
 
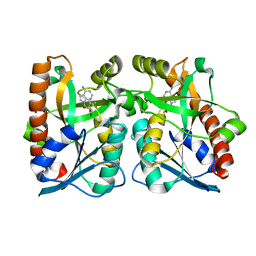 | | Crystal Structure of Arabidopsis thaliana 5'-Methylthioadenosine nucleosidase in complex with 5'-methylthiotubercidin | | Descriptor: | 1,2-ETHANEDIOL, 2-(4-AMINO-PYRROLO[2,3-D]PYRIMIDIN-7-YL)-5-METHYLSULFANYLMETHYL-TETRAHYDRO-FURAN-3,4-DIOL, 5'-methylthioadenosine nucleosidase | | Authors: | Siu, K.K.W, Howell, P.L. | | Deposit date: | 2007-08-02 | | Release date: | 2008-04-01 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.84 Å) | | Cite: | Molecular determinants of substrate specificity in plant 5'-methylthioadenosine nucleosidases.
J.Mol.Biol., 378, 2008
|
|
2QTT
 
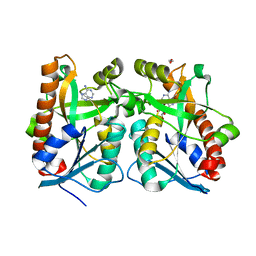 | | Crystal Structure of Arabidopsis thaliana 5'-Methylthioadenosine nucleosidase in complex with Formycin A | | Descriptor: | (1S)-1-(7-amino-1H-pyrazolo[4,3-d]pyrimidin-3-yl)-1,4-anhydro-D-ribitol, 1,2-ETHANEDIOL, 5'-methylthioadenosine nucleosidase, ... | | Authors: | Siu, K.K.W, Howell, P.L. | | Deposit date: | 2007-08-02 | | Release date: | 2008-04-01 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | Molecular determinants of substrate specificity in plant 5'-methylthioadenosine nucleosidases.
J.Mol.Biol., 378, 2008
|
|
2R9C
 
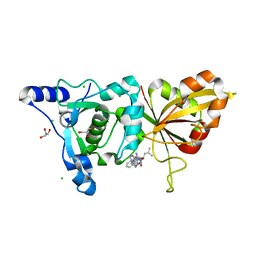 | | Calpain 1 proteolytic core inactivated by ZLAK-3001, an alpha-ketoamide | | Descriptor: | CALCIUM ION, CHLORIDE ION, Calpain-1 catalytic subunit, ... | | Authors: | Qian, J, Campbell, R.L, Davies, P.L. | | Deposit date: | 2007-09-12 | | Release date: | 2008-08-26 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Cocrystal structures of primed side-extending alpha-ketoamide inhibitors reveal novel calpain-inhibitor aromatic interactions.
J.Med.Chem., 51, 2008
|
|
2R9F
 
 | | Calpain 1 proteolytic core inactivated by ZLAK-3002, an alpha-ketoamide | | Descriptor: | CALCIUM ION, CHLORIDE ION, Calpain-1 catalytic subunit, ... | | Authors: | Qian, J, Campbell, R.L, Davies, P.L. | | Deposit date: | 2007-09-12 | | Release date: | 2008-08-26 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Cocrystal structures of primed side-extending alpha-ketoamide inhibitors reveal novel calpain-inhibitor aromatic interactions.
J.Med.Chem., 51, 2008
|
|
2RI9
 
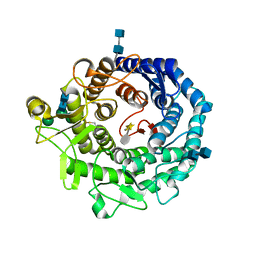 | | Penicillium citrinum alpha-1,2-mannosidase in complex with a substrate analog | | Descriptor: | 2-acetamido-2-deoxy-alpha-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | Authors: | Lobsanov, Y.D, Yoshida, T, Desmet, T, Nerinckx, W, Yip, P, Claeyssens, M, Herscovics, A, Howell, P.L. | | Deposit date: | 2007-10-10 | | Release date: | 2008-03-25 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Modulation of activity by Arg407: structure of a fungal alpha-1,2-mannosidase in complex with a substrate analogue.
Acta Crystallogr.,Sect.D, 64, 2008
|
|
2RI8
 
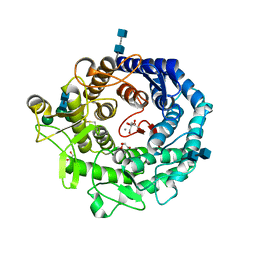 | | Penicillium citrinum alpha-1,2-mannosidase complex with glycerol | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, GLYCEROL, ... | | Authors: | Lobsanov, Y.D, Yoshida, T, Desmet, T, Nerinckx, W, Yip, P, Claeyssens, M, Herscovics, A, Howell, P.L. | | Deposit date: | 2007-10-10 | | Release date: | 2008-03-25 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.16 Å) | | Cite: | Modulation of activity by Arg407: structure of a fungal alpha-1,2-mannosidase in complex with a substrate analogue.
Acta Crystallogr.,Sect.D, 64, 2008
|
|
2W7X
 
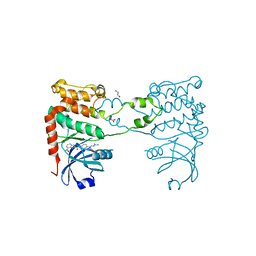 | | Cellular inhibition of checkpoint kinase 2 and potentiation of cytotoxic drugs by novel Chk2 inhibitor PV1019 | | Descriptor: | 1,2-ETHANEDIOL, MAGNESIUM ION, N-[4-[(E)-N-carbamimidamido-C-methyl-carbonimidoyl]phenyl]-7-nitro-1H-indole-2-carboxamide, ... | | Authors: | Jobson, A.G, Lountos, G.T, Lorenzi, P.L, Llamas, J, Connelly, J, Tropea, J.E, Onda, A, Kondapaka, S, Zhang, G, Caplen, N.J, Caredellina, J.H, Monks, A, Self, C, Waugh, D.S, Shoemaker, R.H, Pommier, Y. | | Deposit date: | 2009-01-06 | | Release date: | 2009-09-22 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.07 Å) | | Cite: | Cellular Inhibition of Chk2 Kinase and Potentiation of Camptothecins and Radiation by the Novel Chk2 Inhibitor Pv1019.
J.Pharmacol.Exp.Ther., 331, 2009
|
|
6VE2
 
 | | Tetradecameric PilQ bound by TsaP heptamer from Pseudomonas aeruginosa | | Descriptor: | Fimbrial assembly protein PilQ, LysM domain-containing protein | | Authors: | McCallum, M, Tammam, S, Rubinstein, J.L, Burrows, L.L, Howell, P.L. | | Deposit date: | 2019-12-28 | | Release date: | 2020-12-23 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (4.3 Å) | | Cite: | CryoEM map of Pseudomonas aeruginosa PilQ enables structural characterization of TsaP.
Structure, 29, 2021
|
|
6VE3
 
 | | Tetradecameric PilQ from Pseudomonas aeruginosa | | Descriptor: | Fimbrial assembly protein PilQ | | Authors: | McCallum, M, Tammam, S, Rubinstein, J.L, Burrows, L.L, Howell, P.L. | | Deposit date: | 2019-12-28 | | Release date: | 2020-12-23 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (4.3 Å) | | Cite: | CryoEM map of Pseudomonas aeruginosa PilQ enables structural characterization of TsaP.
Structure, 29, 2021
|
|
