6GS4
 
 | | Crystal structure of peptide transporter DtpA-nanobody in complex with valganciclovir | | Descriptor: | DODECYL-BETA-D-MALTOSIDE, Dipeptide and tripeptide permease A, [(2~{S})-2-[(2-azanyl-6-oxidanylidene-3~{H}-purin-9-yl)methoxy]-3-oxidanyl-propyl] (2~{S})-2-azanyl-3-methyl-butanoate, ... | | Authors: | Ural-Blimke, Y, Flayhan, A, Quistgaard, E.M, Loew, C. | | Deposit date: | 2018-06-13 | | Release date: | 2019-01-30 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.645 Å) | | Cite: | Structure of Prototypic Peptide Transporter DtpA from E. coli in Complex with Valganciclovir Provides Insights into Drug Binding of Human PepT1.
J. Am. Chem. Soc., 141, 2019
|
|
2GSP
 
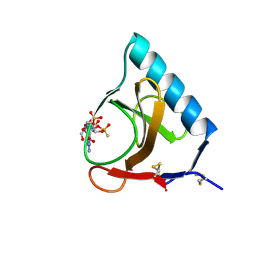 | | RIBONUCLEASE T1/2',3'-CGPS AND 3'-GMP, 2 DAYS | | Descriptor: | CALCIUM ION, GUANOSINE-2',3'-CYCLOPHOSPHOROTHIOATE, GUANOSINE-3'-MONOPHOSPHATE, ... | | Authors: | Zegers, I, Wyns, L. | | Deposit date: | 1997-12-02 | | Release date: | 1998-08-12 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Hydrolysis of a slow cyclic thiophosphate substrate of RNase T1 analyzed by time-resolved crystallography.
Nat.Struct.Biol., 5, 1998
|
|
6F2G
 
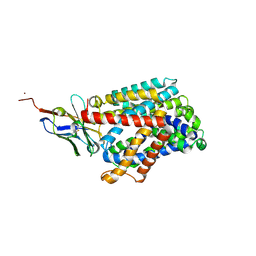 | | Bacterial asc transporter crystal structure in open to in conformation | | Descriptor: | Nanobody 74, Putative amino acid/polyamine transport protein, ZINC ION | | Authors: | Fort, J, Errasti-Murugarren, E, Carpena, X, Palacin, M, Fita, I. | | Deposit date: | 2017-11-24 | | Release date: | 2019-04-24 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.92 Å) | | Cite: | L amino acid transporter structure and molecular bases for the asymmetry of substrate interaction.
Nat Commun, 10, 2019
|
|
6F2W
 
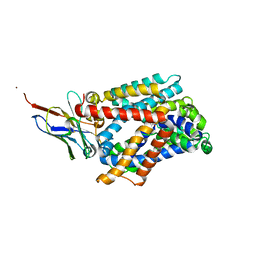 | | Bacterial asc transporter crystal structure in open to in conformation | | Descriptor: | ALPHA-AMINOISOBUTYRIC ACID, Nanobody 74, Putative amino acid/polyamine transport protein, ... | | Authors: | Fort, J, Errasti-Murugarren, E, Carpena, X, Palacin, M, Fita, I. | | Deposit date: | 2017-11-27 | | Release date: | 2019-04-24 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | L amino acid transporter structure and molecular bases for the asymmetry of substrate interaction.
Nat Commun, 10, 2019
|
|
8OUD
 
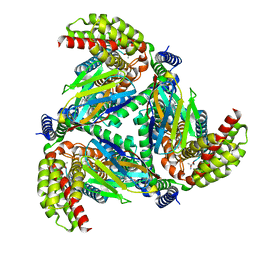 | |
6HLU
 
 | |
6ZG3
 
 | | the structure of ECF PanT transporter in a complex with a nanobody | | Descriptor: | CA14381 nanobody, CITRIC ACID, Conserved hypothetical membrane protein, ... | | Authors: | Setyawati, I, Guskov, A, Slotboom, D.J. | | Deposit date: | 2020-06-18 | | Release date: | 2021-03-03 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | In vitro reconstitution of dynamically interacting integral membrane subunits of energy-coupling factor transporters.
Elife, 9, 2020
|
|
5M7M
 
 | | Novel Imidazo[1,2-a]pyridine Derivatives with Potent Autotaxin/ENPP2 Inhibitor Activity | | Descriptor: | CHLORIDE ION, Ectonucleotide pyrophosphatase/phosphodiesterase family member 2, IODIDE ION, ... | | Authors: | Wolhkoning, A, Fleury, D, Leonard, P, Triballeau, N, Mollat, P, Vercheval, L. | | Deposit date: | 2016-10-28 | | Release date: | 2017-08-30 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Discovery, Structure-Activity Relationship, and Binding Mode of an Imidazo[1,2-a]pyridine Series of Autotaxin Inhibitors.
J. Med. Chem., 60, 2017
|
|
7APJ
 
 | | Structure of autoinhibited Akt1 reveals mechanism of PIP3-mediated activation | | Descriptor: | NB41, RAC-alpha serine/threonine-protein kinase,Non-specific serine/threonine protein kinase,RAC-alpha serine/threonine-protein kinase | | Authors: | Truebestein, L, Hornegger, H, Leonard, T.A. | | Deposit date: | 2020-10-16 | | Release date: | 2021-08-25 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Structure of autoinhibited Akt1 reveals mechanism of PIP 3 -mediated activation.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
5IKC
 
 | | X-RAY STRUCTURE OF THE N-TERMINAL DOMAIN OF HUMAN DOUBLECORTIN in complex with FAB | | Descriptor: | CHLORIDE ION, Ighg protein, MAb 6H10 light chain, ... | | Authors: | Ruf, A, Stihle, M, Benz, J, Thoma, R, Rudolph, M.G. | | Deposit date: | 2016-03-03 | | Release date: | 2016-05-18 | | Last modified: | 2020-03-11 | | Method: | X-RAY DIFFRACTION (2.06 Å) | | Cite: | Crystal Structures of the Human Doublecortin C- and N-terminal Domains in Complex with Specific Antibodies.
J.Biol.Chem., 291, 2016
|
|
6HER
 
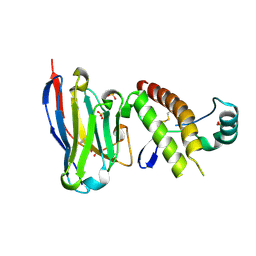 | | Mouse prion protein in complex with Nanobody 484 | | Descriptor: | GLYCEROL, Major prion protein, Nanobody 484, ... | | Authors: | Soror, S.H, Abskharon, R, Wohlkonig, A. | | Deposit date: | 2018-08-20 | | Release date: | 2019-12-04 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.199 Å) | | Cite: | Structural evidence for the critical role of the prion protein hydrophobic region in forming an infectious prion.
Plos Pathog., 15, 2019
|
|
5USF
 
 | |
6HHD
 
 | | Mouse Prion Protein in complex with Nanobody 484 | | Descriptor: | CHLORIDE ION, GLYCEROL, Major prion protein, ... | | Authors: | Soror, S, Abskharon, R, Wohlkonig, A. | | Deposit date: | 2018-08-28 | | Release date: | 2019-12-11 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.102 Å) | | Cite: | Structural evidence for the critical role of the prion protein hydrophobic region in forming an infectious prion.
Plos Pathog., 15, 2019
|
|
5GXB
 
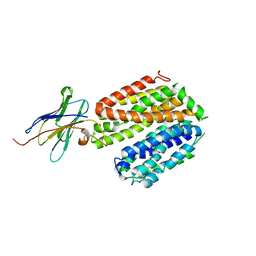 | | crystal structure of a LacY/Nanobody complex | | Descriptor: | Lactose permease, nanobody | | Authors: | Jiang, X, Wu, J.P, Yan, N, Kaback, H.R. | | Deposit date: | 2016-09-16 | | Release date: | 2016-10-26 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Crystal structure of a LacY-nanobody complex in a periplasmic-open conformation.
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
7Q6Z
 
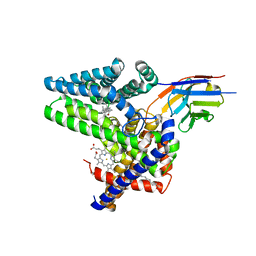 | | Structure of Hedgehog acyltransferase (HHAT) in complex with megabody 177 bound to IMP-1575 | | Descriptor: | 2-(2-methylpropylamino)-1-[(4R)-4-(6-methylpyridin-2-yl)-6,7-dihydro-4H-thieno[3,2-c]pyridin-5-yl]ethanone, CHOLESTEROL, Megabody 177, ... | | Authors: | Coupland, C, Carrique, L, Siebold, C. | | Deposit date: | 2021-11-09 | | Release date: | 2022-01-26 | | Method: | ELECTRON MICROSCOPY (3.59 Å) | | Cite: | Structure, mechanism, and inhibition of Hedgehog acyltransferase.
Mol.Cell, 81, 2021
|
|
5DA0
 
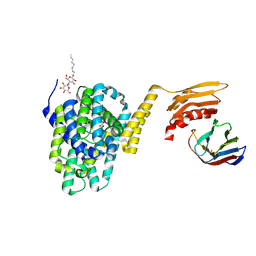 | | Structure of the the SLC26 transporter SLC26Dg in complex with a nanobody | | Descriptor: | DECYL-BETA-D-MALTOPYRANOSIDE, Nanobody, Sulphate transporter | | Authors: | Dutzler, R, Geertsma, E.R, Chang, Y, Shaik, F.R. | | Deposit date: | 2015-08-19 | | Release date: | 2015-09-09 | | Last modified: | 2015-10-14 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Structure of a prokaryotic fumarate transporter reveals the architecture of the SLC26 family.
Nat.Struct.Mol.Biol., 22, 2015
|
|
5DFZ
 
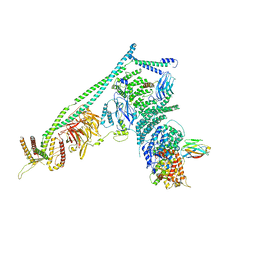 | | Structure of Vps34 complex II from S. cerevisiae. | | Descriptor: | Nanobody binding S. cerevisiae Vps34, Phosphatidylinositol 3-kinase VPS34, Putative N-terminal domain of S. cerevisiae Vps30, ... | | Authors: | Rostislavleva, K, Soler, N, Ohashi, Y, Zhang, L, Williams, R.L. | | Deposit date: | 2015-08-27 | | Release date: | 2015-10-07 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (4.4 Å) | | Cite: | Structure and flexibility of the endosomal Vps34 complex reveals the basis of its function on membranes.
Science, 350, 2015
|
|
5DA4
 
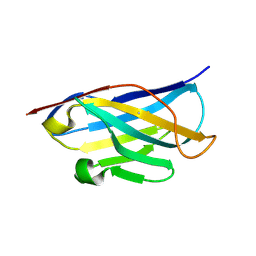 | |
7Q1U
 
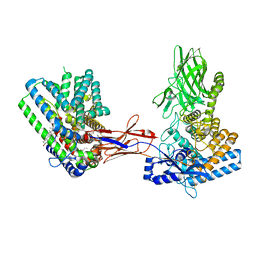 | | Structure of Hedgehog acyltransferase (HHAT) in complex with megabody 177 bound to non-hydrolysable palmitoyl-CoA (Composite Map) | | Descriptor: | CHOLESTEROL, MAGNESIUM ION, Megabody 177, ... | | Authors: | Coupland, C, Carrique, L, Siebold, C. | | Deposit date: | 2021-10-21 | | Release date: | 2022-06-08 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | Structure, mechanism, and inhibition of Hedgehog acyltransferase.
Mol.Cell, 81, 2021
|
|
7PQQ
 
 | |
7PQG
 
 | |
4I73
 
 | | Crystal structure of the Trypanosoma brucei Inosine-Adenosine-Guanosine nucleoside hydrolase in complex with compound UAMC-00312 | | Descriptor: | (2R,3R,4S)-2-(hydroxymethyl)-1-[(4-hydroxythieno[3,2-d]pyrimidin-7-yl)methyl]pyrrolidine-3,4-diol, CALCIUM ION, Inosine-adenosine-guanosine-nucleoside hydrolase, ... | | Authors: | Giannese, F, Degano, M. | | Deposit date: | 2012-11-30 | | Release date: | 2013-08-07 | | Last modified: | 2013-12-18 | | Method: | X-RAY DIFFRACTION (2.18 Å) | | Cite: | Structures of purine nucleosidase from Trypanosoma brucei bound to isozyme-specific trypanocidals and a novel metalorganic inhibitor
Acta Crystallogr.,Sect.D, 69, 2013
|
|
4I71
 
 | | Crystal structure of the Trypanosoma brucei Inosine-Adenosine-Guanosine nucleoside hydrolase in complex with a trypanocidal compound | | Descriptor: | (2R,3R,4S)-1-[(4-amino-5H-pyrrolo[3,2-d]pyrimidin-7-yl)methyl]-2-(hydroxymethyl)pyrrolidine-3,4-diol, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CALCIUM ION, ... | | Authors: | Giannese, F, Degano, M. | | Deposit date: | 2012-11-30 | | Release date: | 2013-08-07 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.28 Å) | | Cite: | Structures of purine nucleosidase from Trypanosoma brucei bound to isozyme-specific trypanocidals and a novel metalorganic inhibitor
Acta Crystallogr.,Sect.D, 69, 2013
|
|
4I72
 
 | | Crystal structure of the Trypanosoma brucei Inosine-Adenosine-Guanosine nucleoside hydrolase in complex with Immucillin A | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 3,4-PYRROLIDINEDIOL,2-(4-AMINO-5H-PYRROLO[3,2-D]PYRIMIDIN-7-YL)-5-(HYDROXYMETHYL)-2S,3S,4R,5R, CALCIUM ION, ... | | Authors: | Giannese, F, Degano, M. | | Deposit date: | 2012-11-30 | | Release date: | 2013-08-07 | | Last modified: | 2013-12-18 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Structures of purine nucleosidase from Trypanosoma brucei bound to isozyme-specific trypanocidals and a novel metalorganic inhibitor
Acta Crystallogr.,Sect.D, 69, 2013
|
|
4I75
 
 | | Crystal structure of the Trypanosoma brucei Inosine-Adenosine-Guanosine nucleoside hydrolase in complex with the NiTris metalorganic complex | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CALCIUM ION, Inosine-adenosine-guanosine-nucleoside hydrolase, ... | | Authors: | Giannese, F, Degano, M. | | Deposit date: | 2012-11-30 | | Release date: | 2013-08-07 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structures of purine nucleosidase from Trypanosoma brucei bound to isozyme-specific trypanocidals and a novel metalorganic inhibitor
Acta Crystallogr.,Sect.D, 69, 2013
|
|
