6UX2
 
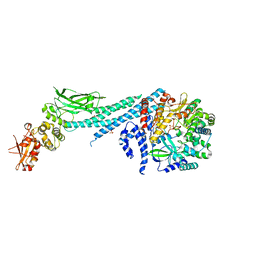 | | Crystal structure of ZIKV RdRp in complex with STAT2 | | Descriptor: | Nonstructural Protein 5, SULFATE ION, Signal transducer and activator of transcription 2, ... | | Authors: | Wang, B, Song, J. | | Deposit date: | 2019-11-06 | | Release date: | 2020-07-08 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (3.01 Å) | | Cite: | Structural basis for STAT2 suppression by flavivirus NS5.
Nat.Struct.Mol.Biol., 27, 2020
|
|
6WCZ
 
 | | CryoEM structure of full-length ZIKV NS5-hSTAT2 complex | | Descriptor: | Non-structural protein 5, Signal transducer and activator of transcription 2, ZINC ION | | Authors: | Boxiao, W, Stephanie, T, Kang, Z, Maria, T.S, Jian, F, Jiuwei, L, Linfeng, G, Wendan, R, Yanxiang, C, Ethan, C.V, HeaJin, H, Matthew, J.E, Sean, E.O, Adolfo, G.S, Hong, Z, Rong, H, Jikui, S. | | Deposit date: | 2020-03-31 | | Release date: | 2020-07-08 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | Structural basis for STAT2 suppression by flavivirus NS5.
Nat.Struct.Mol.Biol., 27, 2020
|
|
2KA4
 
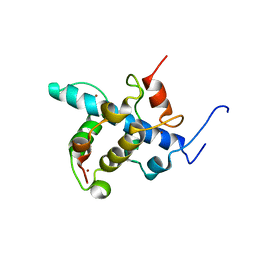 | | NMR structure of the CBP-TAZ1/STAT2-TAD complex | | Descriptor: | Crebbp protein, Signal transducer and activator of transcription 2, ZINC ION | | Authors: | Wojciak, J.M, Martinez-Yamout, M.A, Dyson, H.J, Wright, P.E. | | Deposit date: | 2008-10-30 | | Release date: | 2009-04-21 | | Last modified: | 2024-05-08 | | Method: | SOLUTION NMR | | Cite: | Structural basis for recruitment of CBP/p300 coactivators by STAT1 and STAT2 transactivation domains
Embo J., 28, 2009
|
|
8T12
 
 | |
8T13
 
 | |
5OEN
 
 | | Crystal Structure of STAT2 in complex with IRF9 | | Descriptor: | Interferon regulatory factor 9, Signal transducer and activator of transcription | | Authors: | Rengachari, S, Panne, D. | | Deposit date: | 2017-07-09 | | Release date: | 2018-01-24 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.919 Å) | | Cite: | Structural basis of STAT2 recognition by IRF9 reveals molecular insights into ISGF3 function.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
7ZN7
 
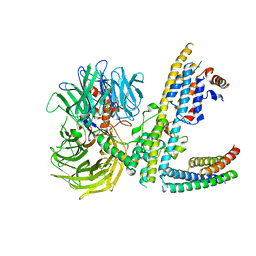 | | Cryo-EM structure of RCMV-E E27 bound to human DDB1 (deltaBPB) and rat STAT2 CCD | | Descriptor: | B27a, DNA damage-binding protein 1, Signal transducer and activator of transcription, ... | | Authors: | Lauer, S, Spahn, C.M.T, Schwefel, D. | | Deposit date: | 2022-04-20 | | Release date: | 2022-11-09 | | Last modified: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (3.78 Å) | | Cite: | Structural mechanism of CRL4-instructed STAT2 degradation via a novel cytomegaloviral DCAF receptor.
Embo J., 42, 2023
|
|
7ZNN
 
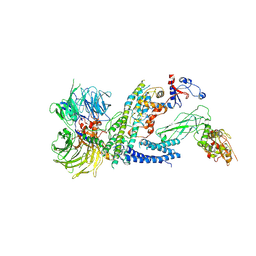 | | Cryo-EM structure of RCMV-E E27 bound to human DDB1 (deltaBPB) and full-length rat STAT2 | | Descriptor: | B27a, DNA damage-binding protein 1, Signal transducer and activator of transcription, ... | | Authors: | Lauer, S, Spahn, C.M.T, Schwefel, D. | | Deposit date: | 2022-04-21 | | Release date: | 2022-11-09 | | Last modified: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (4.8 Å) | | Cite: | Structural mechanism of CRL4-instructed STAT2 degradation via a novel cytomegaloviral DCAF receptor.
Embo J., 42, 2023
|
|
5OEM
 
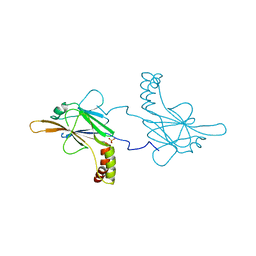 | |
2KA6
 
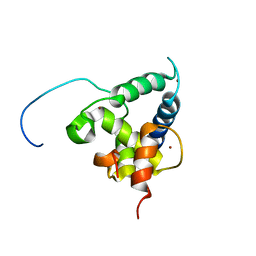 | | NMR structure of the CBP-TAZ2/STAT1-TAD complex | | Descriptor: | CREB-binding protein, Signal transducer and activator of transcription 1-alpha/beta, ZINC ION | | Authors: | Wojciak, J.M, Martinez-Yamout, M.A, Dyson, H.J, Wright, P.E. | | Deposit date: | 2008-10-30 | | Release date: | 2009-04-07 | | Last modified: | 2024-05-08 | | Method: | SOLUTION NMR | | Cite: | Structural basis for recruitment of CBP/p300 coactivators by STAT1 and STAT2 transactivation domains.
Embo J., 28, 2009
|
|
2ASQ
 
 | | Solution Structure of SUMO-1 in Complex with a SUMO-binding Motif (SBM) | | Descriptor: | Protein inhibitor of activated STAT2, Small ubiquitin-related modifier 1 | | Authors: | Song, J, Zhang, Z, Hu, W, Chen, Y. | | Deposit date: | 2005-08-23 | | Release date: | 2005-10-11 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Small Ubiquitin-like Modifier (SUMO) Recognition of a SUMO Binding Motif: A reversal of the bound orientation
J.Biol.Chem., 280, 2005
|
|
7UKN
 
 | | Crystal Structure of DDB1 in Complex with the H-Box Motif of pUL145 | | Descriptor: | DNA damage-binding protein 1, H-Box Motif of pUL145 | | Authors: | Wick, E.T, Treadway, C.J, Nicely, N.I, Li, Z, Ren, Z, Baldwin, A.S, Xiong, Y, Harrison, J.S, Brown, N.G. | | Deposit date: | 2022-04-01 | | Release date: | 2022-08-10 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Insight into Viral Hijacking of CRL4 Ubiquitin Ligase through Structural Analysis of the pUL145-DDB1 Complex.
J.Virol., 96, 2022
|
|
8GZR
 
 | | Cryo-EM structure of the the NS5-NS3 RNA-elongation complex | | Descriptor: | CYTIDINE-5'-DIPHOSPHATE, Genome polyprotein, MANGANESE (II) ION, ... | | Authors: | Osawa, T, Ehara, H, Sekine, S. | | Deposit date: | 2022-09-27 | | Release date: | 2023-08-02 | | Last modified: | 2023-08-23 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Structures of dengue virus RNA replicase complexes.
Mol.Cell, 83, 2023
|
|
8GZQ
 
 | | Cryo-EM structure of the NS5-NS3-SLA complex | | Descriptor: | GUANOSINE-5'-DIPHOSPHATE, Genome polyprotein, RNA, ... | | Authors: | Osawa, T, Ehara, H, Sekine, S. | | Deposit date: | 2022-09-27 | | Release date: | 2023-08-02 | | Last modified: | 2023-08-23 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Structures of dengue virus RNA replicase complexes.
Mol.Cell, 83, 2023
|
|
8GZP
 
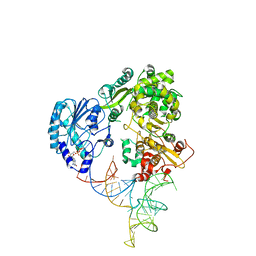 | | Cryo-EM structure of the NS5-SLA complex | | Descriptor: | GUANOSINE-5'-DIPHOSPHATE, Genome polyprotein, MAGNESIUM ION, ... | | Authors: | Osawa, T, Ehara, H, Sekine, S. | | Deposit date: | 2022-09-27 | | Release date: | 2023-08-02 | | Last modified: | 2023-08-23 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Structures of dengue virus RNA replicase complexes.
Mol.Cell, 83, 2023
|
|
3WWT
 
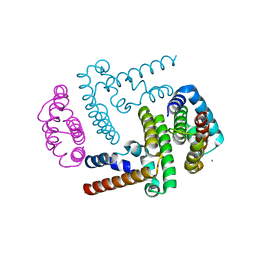 | | Crystal Structure of the Y3:STAT1ND complex | | Descriptor: | C' protein, CALCIUM ION, Signal transducer and activator of transcription 1-alpha/beta | | Authors: | Oda, K, Sakaguchi, T, Matoba, Y. | | Deposit date: | 2014-06-27 | | Release date: | 2015-07-01 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural Basis of the Inhibition of STAT1 Activity by Sendai Virus C Protein.
J.Virol., 89, 2015
|
|
8CXI
 
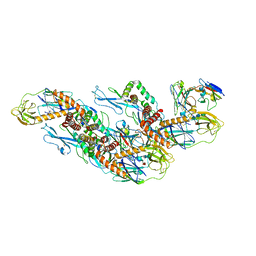 | | Structures of Zika Virus in Complex with Antibodies Targeting E Dimer Epitopes and Basis for Neutralization Efficacy | | Descriptor: | A11 heavy chain, A11 light chain, Ankyrin repeat family A protein 2,Envelope E protein, ... | | Authors: | Liu, W, Zhang, X.K, Gong, D.Y, Dai, X.H, Sharma, A, Zhang, T.H, Rey, F, Zhou, Z.H. | | Deposit date: | 2022-05-21 | | Release date: | 2023-06-07 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Structures of Zika Virus in Complex with Antibodies Targeting E Dimer Epitopes and Basis for Neutralization Efficacy
To Be Published
|
|
3HHC
 
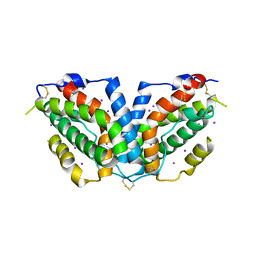 | |
2WZL
 
 | | The Structure of the N-RNA Binding Domain of the Mokola virus Phosphoprotein | | Descriptor: | GLYCEROL, PHOSPHOPROTEIN | | Authors: | Assenberg, R, Delmas, O, Ren, J, Vidalain, P, Verma, A, Larrous, F, Graham, S, Tangy, F, Grimes, J, Bourhy, H. | | Deposit date: | 2009-11-30 | | Release date: | 2009-12-15 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | The Structure of the N-RNA Binding Domain of the Mokola Virus Phosphoprotein
J.Virol., 84, 2010
|
|
5VJ2
 
 | | Structure of human respiratory syncytial virus non-structural protein 1 (NS1) | | Descriptor: | 1,4-DIETHYLENE DIOXIDE, CHLORIDE ION, GLYCEROL, ... | | Authors: | Leung, D.W, Borek, D.M, Amarasinghe, G.K. | | Deposit date: | 2017-04-18 | | Release date: | 2017-07-12 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.22 Å) | | Cite: | Structural basis for human respiratory syncytial virus NS1-mediated modulation of host responses.
Nat Microbiol, 2, 2017
|
|
7F60
 
 | |
4FO9
 
 | | Crystal structure of the E3 SUMO Ligase PIAS2 | | Descriptor: | E3 SUMO-protein ligase PIAS2, UNKNOWN ATOM OR ION, ZINC ION | | Authors: | Dong, A, Hu, J, Dobrovetsky, E, Tempel, W, Bountra, C, Arrowsmith, C.H, Edwards, A.M, Tong, Y, Structural Genomics Consortium (SGC) | | Deposit date: | 2012-06-20 | | Release date: | 2012-07-18 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.39 Å) | | Cite: | Crystal structure of the E3 SUMO Ligase PIAS2
to be published
|
|
7LDK
 
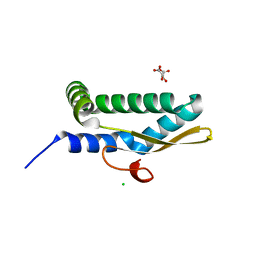 | | Structure of human respiratory syncytial virus nonstructural protein 2 (NS2) | | Descriptor: | CHLORIDE ION, D(-)-TARTARIC ACID, Non-structural protein 2 | | Authors: | Chatterjee, S, Borek, D, Otwinowski, Z, Amarasinghe, G.K, Leung, D.W. | | Deposit date: | 2021-01-13 | | Release date: | 2021-03-17 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.82 Å) | | Cite: | Structural basis for IFN antagonism by human respiratory syncytial virus nonstructural protein 2.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
2HYE
 
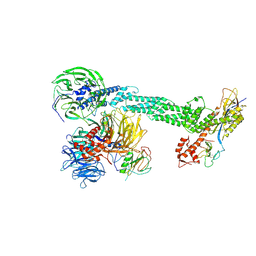 | | Crystal Structure of the DDB1-Cul4A-Rbx1-SV5V Complex | | Descriptor: | Cullin-4A, DNA damage-binding protein 1, Nonstructural protein V, ... | | Authors: | Angers, S, Li, T, Yi, X, MacCoss, M.J, Moon, R.T, Zheng, N. | | Deposit date: | 2006-08-05 | | Release date: | 2006-10-03 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Molecular architecture and assembly of the DDB1-CUL4A ubiquitin ligase machinery.
Nature, 443, 2006
|
|
2B5L
 
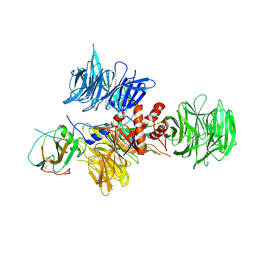 | | Crystal Structure of DDB1 In Complex with Simian Virus 5 V Protein | | Descriptor: | Nonstructural protein V, ZINC ION, damage-specific DNA binding protein 1 | | Authors: | Li, T, Chen, X, Garbutt, K.C, Zhou, P, Zheng, N. | | Deposit date: | 2005-09-28 | | Release date: | 2006-02-28 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Structure of DDB1 in complex with a paramyxovirus V protein: viral hijack of a propeller cluster in ubiquitin ligase.
Cell(Cambridge,Mass.), 124, 2006
|
|
