5ECJ
 
 | |
6BW4
 
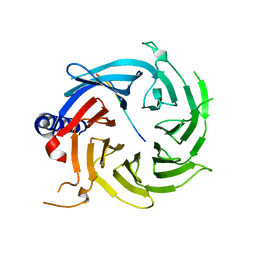 | | Crystal structure of RBBP4 in complex with PRDM16 N-terminal peptide | | Descriptor: | Histone-binding protein RBBP4, PR domain zinc finger protein 16, UNKNOWN ATOM OR ION | | Authors: | Ivanochko, D, Halabelian, L, Hutchinson, A, Seitova, A, Bountra, C, Edwards, A.M, Arrowsmith, C.H, Structural Genomics Consortium (SGC) | | Deposit date: | 2017-12-14 | | Release date: | 2017-12-27 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Direct interaction between the PRDM3 and PRDM16 tumor suppressors and the NuRD chromatin remodeling complex.
Nucleic Acids Res., 47, 2019
|
|
2N1I
 
 | |
6BW3
 
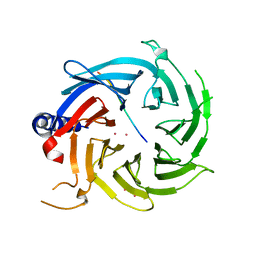 | | Crystal structure of RBBP4 in complex with PRDM3 N-terminal peptide | | Descriptor: | Histone-binding protein RBBP4, MDS1 and EVI1 complex locus protein MDS1, UNKNOWN ATOM OR ION | | Authors: | Ivanochko, D, Halabelian, L, Hutchinson, A, Seitova, A, Bountra, C, Edwards, A.M, Arrowsmith, C.H, Structural Genomics Consortium (SGC) | | Deposit date: | 2017-12-14 | | Release date: | 2017-12-27 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Direct interaction between the PRDM3 and PRDM16 tumor suppressors and the NuRD chromatin remodeling complex.
Nucleic Acids Res., 47, 2019
|
|
3DAL
 
 | | Methyltransferase domain of human PR domain-containing protein 1 | | Descriptor: | PR domain zinc finger protein 1 | | Authors: | Amaya, M.F, Zeng, H, Antoshenko, T, Dong, A, Loppnau, P, Bountra, C, Weigelt, J, Arrowsmith, C.H, Edwards, A.M, Bochkarev, A, Min, J, Plotnikov, A.N, Wu, H, Structural Genomics Consortium (SGC) | | Deposit date: | 2008-05-29 | | Release date: | 2008-08-12 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | The crystal structure
of methyltransferase domain of human PR domain-containing protein 1
To be Published
|
|
3IHX
 
 | | Methyltransferase domain of human PR domain-containing protein 10 | | Descriptor: | PR domain zinc finger protein 10 | | Authors: | Amaya, M.F, Dombrovski, L, Ni, S, Bountra, C, Weigelt, J, Arrowsmith, C.H, Edwards, A.M, Botchkarev, A, Min, J, Plotnikov, A.N, Wu, H, Structural Genomics Consortium (SGC) | | Deposit date: | 2009-07-31 | | Release date: | 2009-08-18 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Methyltransferase domain of human PR domain-containing protein 10
To be Published
|
|
3EP0
 
 | | Methyltransferase domain of human PR domain-containing protein 12 | | Descriptor: | PR domain zinc finger protein 12 | | Authors: | Amaya, M.F, Zeng, H, Loppnau, P, Bountra, C, Weigelt, J, Arrowsmith, C.H, Edwards, A.M, Botchkarev, A, Min, J, Plotnikov, A.N, Wu, H, Structural Genomics Consortium (SGC) | | Deposit date: | 2008-09-29 | | Release date: | 2008-11-04 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | The crystal structure of methyltransferase domain of human PR
Domain-containing protein 12.
To be Published
|
|
3RAY
 
 | | Crystal structure of Methyltransferase domain of human PR domain-containing protein 11 | | Descriptor: | CHLORIDE ION, PR domain-containing protein 11, SODIUM ION, ... | | Authors: | Dong, A, Zeng, H, Loppnau, P, Walker, J.R, Bountra, C, Weigelt, J, Arrowsmith, C.H, Edwards, A.M, Min, J, Wu, H, Structural Genomics Consortium (SGC) | | Deposit date: | 2011-03-28 | | Release date: | 2011-04-20 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.73 Å) | | Cite: | Crystal structure of Methyltransferase domain of human PR domain-containing protein 11
To be Published
|
|
1QZZ
 
 | | Crystal structure of aclacinomycin-10-hydroxylase (RdmB) in complex with S-adenosyl-L-methionine (SAM) | | Descriptor: | ACETATE ION, S-ADENOSYLMETHIONINE, aclacinomycin-10-hydroxylase | | Authors: | Jansson, A, Niemi, J, Lindqvist, Y, Mantsala, P, Schneider, G, Structural Proteomics in Europe (SPINE) | | Deposit date: | 2003-09-19 | | Release date: | 2003-11-25 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal Structure of Aclacinomycin-10-Hydroxylase, a S-Adenosyl-L-Methionine-dependent Methyltransferase Homolog Involved in Anthracycline Biosynthesis in Streptomyces purpurascens.
J.Mol.Biol., 334, 2003
|
|
1R00
 
 | | Crystal structure of aclacinomycin-10-hydroxylase (RdmB) in complex with S-adenosyl-L-homocysteine (SAH) | | Descriptor: | ACETATE ION, S-ADENOSYL-L-HOMOCYSTEINE, aclacinomycin-10-hydroxylase | | Authors: | Jansson, A, Niemi, J, Lindqvist, Y, Mantsala, P, Schneider, G. | | Deposit date: | 2003-09-19 | | Release date: | 2003-11-25 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal Structure of Aclacinomycin-10-Hydroxylase, a S-Adenosyl-L-Methionine-dependent Methyltransferase Homolog Involved in Anthracycline Biosynthesis in Streptomyces purpurascens.
J.Mol.Biol., 334, 2003
|
|
1XDS
 
 | | Crystal structure of Aclacinomycin-10-hydroxylase (RdmB) in complex with S-adenosyl-L-methionine (SAM) and 11-deoxy-beta-rhodomycin (DbrA) | | Descriptor: | 11-DEOXY-BETA-RHODOMYCIN, Protein RdmB, S-ADENOSYLMETHIONINE | | Authors: | Jansson, A, Koskiniemi, H, Erola, A, Wang, J, Mantsala, P, Schneider, G, Niemi, J, Structural Proteomics in Europe (SPINE) | | Deposit date: | 2004-09-08 | | Release date: | 2004-11-23 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Aclacinomycin 10-Hydroxylase Is a Novel Substrate-assisted Hydroxylase Requiring S-Adenosyl-L-methionine as Cofactor
J.Biol.Chem., 280, 2005
|
|
1XDU
 
 | | Crystal structure of Aclacinomycin-10-hydroxylase (RdmB) in complex with Sinefungin (SFG) | | Descriptor: | ACETATE ION, Protein RdmB, SINEFUNGIN | | Authors: | Jansson, A, Koskiniemi, H, Erola, A, Wang, J, Mantsala, P, Schneider, G, Niemi, J. | | Deposit date: | 2004-09-08 | | Release date: | 2004-11-23 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Aclacinomycin 10-Hydroxylase Is a Novel Substrate-assisted Hydroxylase Requiring S-Adenosyl-L-methionine as Cofactor
J.Biol.Chem., 280, 2005
|
|
