4US5
 
 | | Crystal Structure of apo-MsnO8 | | Descriptor: | 1-ETHOXY-2-(2-ETHOXYETHOXY)ETHANE, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, CHLORIDE ION, ... | | Authors: | Maier, S, Pflueger, T, Loesgen, S, Asmus, K, Broetz, E, Paululat, T, Zeeck, A, Andrade, S, Bechthold, A. | | Deposit date: | 2014-07-03 | | Release date: | 2014-07-16 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Insights Into the Bioactivity of Mensacarcin and Epoxide Formation by Msno8.
Chembiochem, 15, 2014
|
|
3RAO
 
 | | Crystal Structure of the Luciferase-like Monooxygenase from Bacillus cereus ATCC 10987. | | Descriptor: | Putative Luciferase-like Monooxygenase, SULFATE ION | | Authors: | Domagalski, M.J, Chruszcz, M, Xu, X, Cui, H, Chin, S, Savchenko, A, Edwards, A, Joachimiak, A, Minor, W, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2011-03-28 | | Release date: | 2011-05-11 | | Last modified: | 2022-04-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal Structure of the Luciferase-like Monooxygenase from Bacillus cereus ATCC 10987.
To be Published
|
|
4UWM
 
 | | Type II Baeyer-Villiger monooxygenase.The oxygenating constituent of 3,6-diketocamphane monooxygenase from CAM plasmid of Pseudomonas putida in complex with FMN. | | Descriptor: | 3,6-DIKETOCAMPHANE 1,6 MONOOXYGENASE, DI(HYDROXYETHYL)ETHER, FLAVIN MONONUCLEOTIDE, ... | | Authors: | Isupov, M.N, Schroeder, E, Gibson, R.P, Beecher, J, Donadio, G, Saneei, V, Dcunha, S, McGhie, E.J, Sayer, C, Davenport, C.F, Lau, P.C, Hasegawa, Y, Iwaki, H, Kadow, M, Loschinski, K, Bornscheuer, U.T, Bourenkov, G, Littlechild, J.A. | | Deposit date: | 2014-08-12 | | Release date: | 2015-08-26 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The Oxygenating Constituent of 3,6-Diketocamphane Monooxygenase from the Cam Plasmid of Pseudomonas Putida: The First Crystal Structure of a Type II Baeyer-Villiger Monooxygenase.
Acta Crystallogr.,Sect.D, 71, 2015
|
|
8CBB
 
 | |
4J2P
 
 | |
2I7G
 
 | | Crystal Structure of Monooxygenase from Agrobacterium tumefaciens | | Descriptor: | DI(HYDROXYETHYL)ETHER, Monooxygenase, SULFATE ION | | Authors: | Kim, Y, Xu, X, Zheng, H, Joachimiak, A, Edwards, A, Savchenko, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2006-08-30 | | Release date: | 2006-10-03 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.73 Å) | | Cite: | The Crystal Structure of Monooxygenase from Agrobacterium tumefaciens
To be Published, 2006
|
|
3SDO
 
 | |
5DQP
 
 | | EDTA monooxygenase (EmoA) from Chelativorans sp. BNC1 | | Descriptor: | 2-{2-[2-(2-{2-[2-(2-ETHOXY-ETHOXY)-ETHOXY]-ETHOXY}-ETHOXY)-ETHOXY]-ETHOXY}-ETHANOL, EDTA monooxygenase, SULFATE ION | | Authors: | Jun, S.Y, Youn, B, Xun, L, Kang, C, Lewis, K.M. | | Deposit date: | 2015-09-15 | | Release date: | 2016-03-16 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.146 Å) | | Cite: | Structural and biochemical characterization of EDTA monooxygenase and its physical interaction with a partner flavin reductase.
Mol.Microbiol., 100, 2016
|
|
1BRL
 
 | |
5AEC
 
 | | Type II Baeyer-Villiger monooxygenase.The oxygenating constituent of 3,6-diketocamphane monooxygenase from CAM plasmid of Pseudomonas putida in complex with FMN. | | Descriptor: | 3,6-DIKETOCAMPHANE 1,6 MONOOXYGENASE, CHLORIDE ION, GLYCEROL, ... | | Authors: | Isupov, M.N, Schroeder, E, Gibson, R.P, Beecher, J, Donadio, G, Saneei, V, Dcunha, S, McGhie, E.J, Sayer, C, Davenport, C.F, Lau, P.C, Hasegawa, Y, Iwaki, H, Kadow, M, Loschinski, K, Bornscheuer, U.T, Bourenkov, G, Littlechild, J.A. | | Deposit date: | 2015-08-28 | | Release date: | 2015-09-09 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | The Oxygenating Constituent of 3,6-Diketocamphane Monooxygenase from the Cam Plasmid of Pseudomonas Putida: The First Crystal Structure of a Type II Baeyer-Villiger Monooxygenase.
Acta Crystallogr.,Sect.D, 71, 2015
|
|
5LXE
 
 | | F420-dependent glucose-6-phosphate dehydrogenase from Rhodococcus jostii RHA1 | | Descriptor: | F420-dependent glucose-6-phosphate dehydrogenase 1, GLYCEROL, SULFATE ION | | Authors: | Nguyen, Q.-T, Trinco, G, Binda, C, Mattevi, A, Fraaije, M.W. | | Deposit date: | 2016-09-20 | | Release date: | 2016-12-28 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.47 Å) | | Cite: | Discovery and characterization of an F420-dependent glucose-6-phosphate dehydrogenase (Rh-FGD1) from Rhodococcus jostii RHA1.
Appl. Microbiol. Biotechnol., 101, 2017
|
|
6FRI
 
 | |
6SGN
 
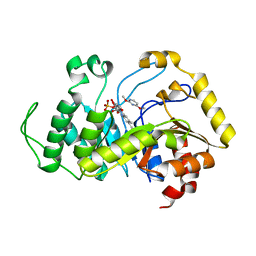 | | Crystal structure of monooxygenase RutA complexed with 2,4-dimethoxypyrimidine. | | Descriptor: | 2,4-dimethoxypyrimidine, FLAVIN MONONUCLEOTIDE, Pyrimidine monooxygenase RutA | | Authors: | Saleem-Batcha, R, Matthews, A, Teufel, R. | | Deposit date: | 2019-08-05 | | Release date: | 2020-02-05 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.501 Å) | | Cite: | Aminoperoxide adducts expand the catalytic repertoire of flavin monooxygenases.
Nat.Chem.Biol., 16, 2020
|
|
2B81
 
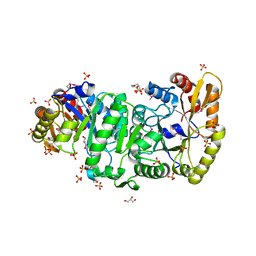 | | Crystal Structure of the Luciferase-like Monooxygenase from Bacillus cereus | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, GLYCEROL, Luciferase-like monooxygenase, ... | | Authors: | Kim, Y, Li, H, Moy, S, Collart, F, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2005-10-06 | | Release date: | 2005-11-22 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal Structure of the Luciferase-like Monooxygenase from Bacillus cereus
To be Published
|
|
1LUC
 
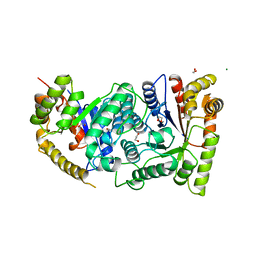 | | BACTERIAL LUCIFERASE | | Descriptor: | 1,2-ETHANEDIOL, BACTERIAL LUCIFERASE, MAGNESIUM ION | | Authors: | Fisher, A.J, Rayment, I. | | Deposit date: | 1996-05-10 | | Release date: | 1996-12-07 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | The 1.5-A resolution crystal structure of bacterial luciferase in low salt conditions.
J.Biol.Chem., 271, 1996
|
|
3C8N
 
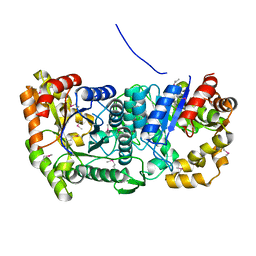 | |
7OH2
 
 | |
7OLF
 
 | |
1RHC
 
 | | F420-dependent secondary alcohol dehydrogenase in complex with an F420-acetone adduct | | Descriptor: | ACETONE, CHLORIDE ION, COENZYME F420, ... | | Authors: | Aufhammer, S.W, Warkentin, E, Berk, H, Shima, S, Thauer, R.K, Ermler, U. | | Deposit date: | 2003-11-14 | | Release date: | 2004-03-30 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Coenzyme binding in f(420)-dependent secondary alcohol dehydrogenase, a member of the bacterial luciferase family.
Structure, 12, 2004
|
|
3FGC
 
 | | Crystal Structure of the Bacterial Luciferase:Flavin Complex Reveals the Basis of Intersubunit Communication | | Descriptor: | Alkanal monooxygenase alpha chain, Alkanal monooxygenase beta chain, FLAVIN MONONUCLEOTIDE, ... | | Authors: | Campbell, Z.T, Weichsel, A, Montfort, W.R, Baldwin, T.O. | | Deposit date: | 2008-12-05 | | Release date: | 2009-05-26 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of the bacterial luciferase/flavin complex provides insight into the function of the beta subunit.
Biochemistry, 48, 2009
|
|
6LR1
 
 | |
1TVL
 
 | |
6KET
 
 | |
8QPL
 
 | | F420-Dependent Methylene-Tetrahydromethanopterin Reductase with F420 from Methanocaldococcus jannaschii | | Descriptor: | 5,10-methylenetetrahydromethanopterin reductase, COENZYME F420 | | Authors: | Gehl, M, Demmer, U, Ermler, U, Shima, S. | | Deposit date: | 2023-10-02 | | Release date: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Mutational and structural studies of ( beta alpha ) 8 -barrel fold methylene-tetrahydropterin reductases utilizing a common catalytic mechanism.
Protein Sci., 33, 2024
|
|
8QPM
 
 | | Structure of methylene-tetrahydromethanopterin reductase from Methanocaldococcus jannaschii | | Descriptor: | 5,10-methylenetetrahydromethanopterin reductase | | Authors: | Gehl, M, Demmer, U, Ermler, U, Shima, S. | | Deposit date: | 2023-10-02 | | Release date: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Mutational and structural studies of ( beta alpha ) 8 -barrel fold methylene-tetrahydropterin reductases utilizing a common catalytic mechanism.
Protein Sci., 33, 2024
|
|
