8R0S
 
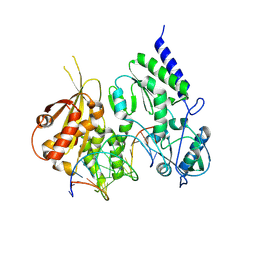 | | Structure of reverse transcriptase from Cauliflower Mosaic Virus in complex with RNA/DNA hybrid | | Descriptor: | DNA (5'-D(*GP*CP*TP*AP*CP*GP*CP*AP*CP*TP*GP*CP*TP*GP*GP*A)-3'), Enzymatic polyprotein, RNA (5'-R(*GP*UP*CP*CP*AP*GP*CP*AP*GP*UP*GP*CP*GP*UP*AP*GP*C)-3') | | Authors: | Prabaharan, C, Figiel, M, Chamera, S, Szczepanowski, R, Nowak, E, Nowotny, M. | | Deposit date: | 2023-10-31 | | Release date: | 2024-07-24 | | Last modified: | 2024-08-21 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Structural and biochemical characterization of cauliflower mosaic virus reverse transcriptase.
J.Biol.Chem., 300, 2024
|
|
8X22
 
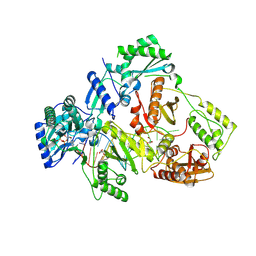 | | HIV-1 reverse transcriptase mutant Q151M/Y115F/F116Y/L74V:DNA:dGTP ternary complex | | Descriptor: | 2'-DEOXYGUANOSINE-5'-TRIPHOSPHATE, DNA/RNA (38-MER), GLYCEROL, ... | | Authors: | Yasutake, Y, Hattori, S.I, Mitsuya, H. | | Deposit date: | 2023-11-09 | | Release date: | 2024-07-17 | | Method: | X-RAY DIFFRACTION (2.31 Å) | | Cite: | Deviated binding of anti-HBV nucleoside analog E-CFCP-TP to the reverse transcriptase active site attenuates the effect of drug-resistant mutations.
Sci Rep, 14, 2024
|
|
8X21
 
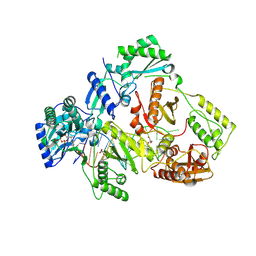 | | HIV-1 reverse transcriptase mutant Q151M/Y115F/F116Y/L74V:DNA:ETV-TP ternary complex | | Descriptor: | DNA/RNA (38-MER), GLYCEROL, HIV-1 RT p51 subunit, ... | | Authors: | Yasutake, Y, Hattori, S.I, Mitsuya, H. | | Deposit date: | 2023-11-09 | | Release date: | 2024-07-17 | | Method: | X-RAY DIFFRACTION (2.33 Å) | | Cite: | Deviated binding of anti-HBV nucleoside analog E-CFCP-TP to the reverse transcriptase active site attenuates the effect of drug-resistant mutations.
Sci Rep, 14, 2024
|
|
8X1Z
 
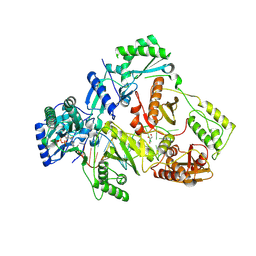 | | HIV-1 reverse transcriptase mutant Q151M/Y115F/F116Y:DNA:E-CFCP-TP ternary complex | | Descriptor: | DNA/RNA (38-MER), E-CFCP-triphosphate, GLYCEROL, ... | | Authors: | Yasutake, Y, Hattori, S.I, Mitsuya, H. | | Deposit date: | 2023-11-09 | | Release date: | 2024-07-17 | | Method: | X-RAY DIFFRACTION (2.62 Å) | | Cite: | Deviated binding of anti-HBV nucleoside analog E-CFCP-TP to the reverse transcriptase active site attenuates the effect of drug-resistant mutations.
Sci Rep, 14, 2024
|
|
8X20
 
 | | HIV-1 reverse transcriptase mutant Q151M/Y115F/F116Y/L74V:DNA:E-CFCP-TP ternary complex | | Descriptor: | DNA/RNA (38-MER), E-CFCP-triphosphate, GLYCEROL, ... | | Authors: | Yasutake, Y, Hattori, S.I, Mitsuya, H. | | Deposit date: | 2023-11-09 | | Release date: | 2024-07-17 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Deviated binding of anti-HBV nucleoside analog E-CFCP-TP to the reverse transcriptase active site attenuates the effect of drug-resistant mutations.
Sci Rep, 14, 2024
|
|
8QBM
 
 | |
8QBL
 
 | |
8QBK
 
 | |
8WUT
 
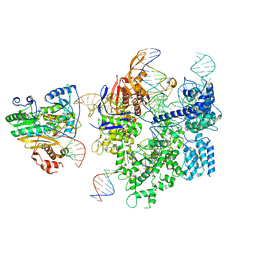 | | SpCas9-MMLV RT-pegRNA-target DNA complex (initiation) | | Descriptor: | CRISPR-associated endonuclease Cas9/Csn1, DNA (5'-D(*TP*GP*AP*TP*GP*GP*CP*AP*GP*AP*GP*TP*AP*CP*TP*AP*G)-3'), DNA (51-MER), ... | | Authors: | Shuto, Y, Nakagawa, R, Hoki, M, Omura, S.N, Hirano, H, Itoh, Y, Nureki, O. | | Deposit date: | 2023-10-21 | | Release date: | 2024-06-05 | | Last modified: | 2024-09-11 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Structural basis for pegRNA-guided reverse transcription by a prime editor.
Nature, 631, 2024
|
|
8WUS
 
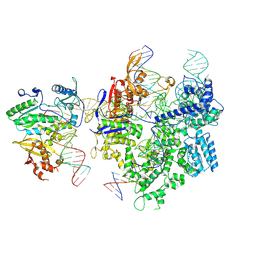 | | SpCas9-MMLV RT-pegRNA-target DNA complex (termination) | | Descriptor: | CRISPR-associated endonuclease Cas9/Csn1, DNA (40-MER), DNA (5'-D(*TP*GP*AP*TP*GP*GP*CP*AP*GP*AP*GP*TP*AP*CP*TP*AP*G)-3'), ... | | Authors: | Shuto, Y, Nakagawa, R, Hoki, M, Omura, S.N, Hirano, H, Itoh, Y, Nureki, O. | | Deposit date: | 2023-10-21 | | Release date: | 2024-06-05 | | Last modified: | 2024-09-11 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Structural basis for pegRNA-guided reverse transcription by a prime editor.
Nature, 631, 2024
|
|
8WUV
 
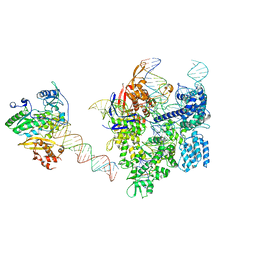 | | SpCas9-MMLV RT-pegRNA-target DNA complex (elongation 16-nt) | | Descriptor: | CRISPR-associated endonuclease Cas9/Csn1, DNA (5'-D(*TP*GP*AP*TP*GP*GP*CP*AP*GP*AP*GP*TP*AP*CP*TP*AP*G)-3'), DNA (50-MER), ... | | Authors: | Shuto, Y, Nakagawa, R, Hoki, M, Omura, S.N, Hirano, H, Itoh, Y, Nureki, O. | | Deposit date: | 2023-10-21 | | Release date: | 2024-06-05 | | Last modified: | 2024-09-11 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Structural basis for pegRNA-guided reverse transcription by a prime editor.
Nature, 631, 2024
|
|
8YGJ
 
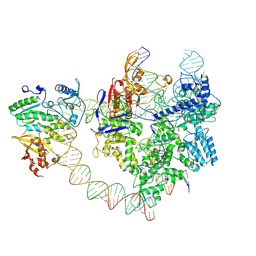 | | SpCas9-MMLV RT-pegRNA-target DNA complex (elongation 28-nt) | | Descriptor: | CRISPR-associated endonuclease Cas9/Csn1, DNA (5'-D(P*TP*GP*AP*TP*GP*GP*CP*AP*GP*AP*GP*TP*AP*CP*TP*AP*G)-3'), DNA (51-MER), ... | | Authors: | Shuto, Y, Nakagawa, R, Hoki, M, Omura, S.N, Hirano, H, Itoh, Y, Nureki, O. | | Deposit date: | 2024-02-26 | | Release date: | 2024-06-05 | | Last modified: | 2024-09-11 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Structural basis for pegRNA-guided reverse transcription by a prime editor.
Nature, 631, 2024
|
|
8SXU
 
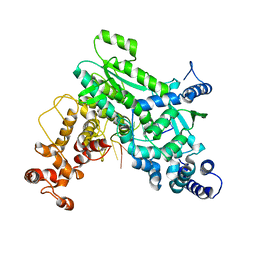 | |
8SXT
 
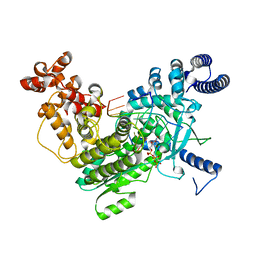 | | Structure of LINE-1 ORF2p with template:primer hybrid | | Descriptor: | DNA primer, LINE-1 retrotransposable element ORF2 protein, MAGNESIUM ION, ... | | Authors: | van Eeuwen, T, Taylor, M.S, Rout, M.P. | | Deposit date: | 2023-05-24 | | Release date: | 2024-01-10 | | Last modified: | 2024-02-14 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Structures, functions and adaptations of the human LINE-1 ORF2 protein.
Nature, 626, 2024
|
|
8C8J
 
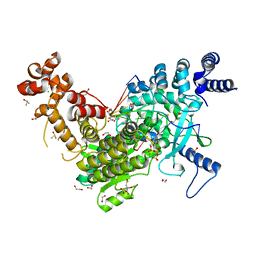 | | Long Interspersed Nuclear Element 1 (LINE-1) reverse transcriptase ternary complex with hybrid duplex and dTTP | | Descriptor: | 1,2-ETHANEDIOL, 1,4-DIETHYLENE DIOXIDE, CHLORIDE ION, ... | | Authors: | Nichols, C.E, Walpole, T.B, Baldwin, E. | | Deposit date: | 2023-01-20 | | Release date: | 2023-12-20 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structures, functions and adaptations of the human LINE-1 ORF2 protein.
Nature, 626, 2024
|
|
8UW3
 
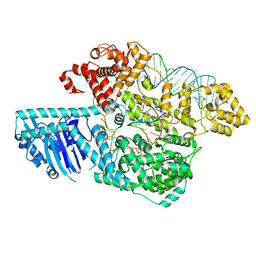 | | Human LINE-1 retrotransposon ORF2 protein engaged with template RNA in elongation state | | Descriptor: | Complementary DNA, LINE-1 retrotransposable element ORF2 protein, THYMIDINE-5'-TRIPHOSPHATE, ... | | Authors: | Thawani, A, Florez Ariza, A.J, Collins, K, Nogales, E. | | Deposit date: | 2023-11-06 | | Release date: | 2023-12-20 | | Last modified: | 2024-02-14 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Template and target-site recognition by human LINE-1 in retrotransposition.
Nature, 626, 2024
|
|
8FLI
 
 | | Cryo-EM structure of a group II intron immediately before branching | | Descriptor: | Group II Intron, MAGNESIUM ION, Maturase reverse transcriptase | | Authors: | Haack, D.B, Rudolfs, B.G, Zhang, C, Lyumkis, D, Toor, N. | | Deposit date: | 2022-12-21 | | Release date: | 2023-12-13 | | Last modified: | 2024-02-07 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Structural basis of branching during RNA splicing.
Nat.Struct.Mol.Biol., 31, 2024
|
|
8STT
 
 | | Crystal Structure of HIV-1 Reverse Transcriptase (Y181C, V106A) varient in Complex with 8-(2-(2-(2,4-dioxo-3,4-dihydropyrimidin-1(2H)-yl)ethoxy)phenoxy)indolizine-2-carbonitrile (JLJ555), a non-nucleoside inhibitor | | Descriptor: | 8-{2-[2-(2,4-dioxo-3,4-dihydropyrimidin-1(2H)-yl)ethoxy]phenoxy}indolizine-2-carbonitrile, MAGNESIUM ION, Reverse transcriptase/ribonuclease H, ... | | Authors: | Hollander, K, Jorgensen, W.L, Anderson, K.S. | | Deposit date: | 2023-05-11 | | Release date: | 2023-11-29 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.62 Å) | | Cite: | Exploring novel HIV-1 reverse transcriptase inhibitors with drug-resistant mutants: A double mutant surprise.
Protein Sci., 32, 2023
|
|
8STP
 
 | | Crystal Structure of HIV-1 Reverse Transcriptase (Y181C) varient in Complex with 8-(2-(2-(2,4-dioxo-3,4-dihydropyrimidin-1(2H)-yl)ethoxy)phenoxy)indolizine-2-carbonitrile (JLJ555), a non-nucleoside inhibitor | | Descriptor: | 8-{2-[2-(2,4-dioxo-3,4-dihydropyrimidin-1(2H)-yl)ethoxy]phenoxy}indolizine-2-carbonitrile, Reverse transcriptase/ribonuclease H, p51 RT | | Authors: | Hollander, K, Frey, K.M, Jorgensen, W.L, Anderson, K.S. | | Deposit date: | 2023-05-11 | | Release date: | 2023-11-29 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (3.09 Å) | | Cite: | Exploring novel HIV-1 reverse transcriptase inhibitors with drug-resistant mutants: A double mutant surprise.
Protein Sci., 32, 2023
|
|
8STR
 
 | | Crystal Structure of HIV-1 Reverse Transcriptase (Y181C) varient in Complex with 5-(2-(2-(2,4-dioxo-3,4-dihydropyrimidin-1(2H)-yl)ethoxy)-4-fluorophenoxy)-7-fluoro-2-naphthonitrile (JLJ636), a non-nucleoside inhibitor | | Descriptor: | 5-{2-[2-(2,4-dioxo-3,4-dihydropyrimidin-1(2H)-yl)ethoxy]-4-fluorophenoxy}-7-fluoronaphthalene-2-carbonitrile, Reverse transcriptase/ribonuclease H, p51 RT | | Authors: | Hollander, K, Chan, A.H, Jorgensen, W.L, Anderson, K.S. | | Deposit date: | 2023-05-11 | | Release date: | 2023-11-29 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.77 Å) | | Cite: | Exploring novel HIV-1 reverse transcriptase inhibitors with drug-resistant mutants: A double mutant surprise.
Protein Sci., 32, 2023
|
|
8STS
 
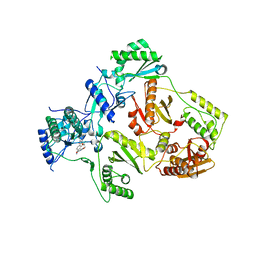 | | Crystal Structure of HIV-1 Reverse Transcriptase (Y181C, V106A) varient in Complex with 5-(2-(2-(2,4-dioxo-3,4-dihydropyrimidin-1(2H)-yl)ethoxy)-4-fluorophenoxy)-7-fluoro-2-naphthonitrile (JLJ636), a non-nucleoside inhibitor | | Descriptor: | 5-{2-[2-(2,4-dioxo-3,4-dihydropyrimidin-1(2H)-yl)ethoxy]-4-fluorophenoxy}-7-fluoronaphthalene-2-carbonitrile, MAGNESIUM ION, Reverse transcriptase/ribonuclease H, ... | | Authors: | Hollander, K, Jorgensen, W.L, Anderson, K.S. | | Deposit date: | 2023-05-11 | | Release date: | 2023-11-29 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (3.02 Å) | | Cite: | Exploring novel HIV-1 reverse transcriptase inhibitors with drug-resistant mutants: A double mutant surprise.
Protein Sci., 32, 2023
|
|
8STU
 
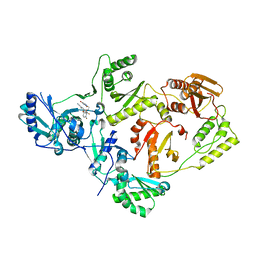 | | Crystal Structure of HIV-1 Reverse Transcriptase (Y181C, V106A) variant in Complex with 8-(2-(2-(2,4-dioxo-3,4-dihydropyrimidin-1(2H)-yl)ethoxy)-4-fluorophenoxy)-6-fluoroindolizine-2-carbonitrile (JLJ578), a non-nucleoside inhibitor | | Descriptor: | 8-{2-[2-(2,4-dioxo-3,4-dihydropyrimidin-1(2H)-yl)ethoxy]-4-fluorophenoxy}-6-fluoroindolizine-2-carbonitrile, MAGNESIUM ION, Reverse transcriptase/ribonuclease H, ... | | Authors: | Hollander, K, Jorgensen, W.L, Anderson, K.S. | | Deposit date: | 2023-05-11 | | Release date: | 2023-11-29 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.76 Å) | | Cite: | Exploring novel HIV-1 reverse transcriptase inhibitors with drug-resistant mutants: A double mutant surprise.
Protein Sci., 32, 2023
|
|
8STV
 
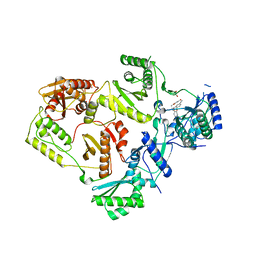 | | Crystal Structure of HIV-1 Reverse Transcriptase (Y181C, V106A) variant in Complex with 5-(2-(2-(2,4-dioxo-3,4-dihydropyrimidin-1(2H)-yl)ethoxy)phenoxy)-2-naphthonitrile (JLJ600), a non-nucleoside inhibitor | | Descriptor: | 1,2-ETHANEDIOL, 5-{2-[2-(2,4-dioxo-3,4-dihydropyrimidin-1(2H)-yl)ethoxy]phenoxy}naphthalene-2-carbonitrile, MAGNESIUM ION, ... | | Authors: | Hollander, K, Jorgensen, W.L, Anderson, K.S. | | Deposit date: | 2023-05-11 | | Release date: | 2023-11-29 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.78 Å) | | Cite: | Exploring novel HIV-1 reverse transcriptase inhibitors with drug-resistant mutants: A double mutant surprise.
Protein Sci., 32, 2023
|
|
8STQ
 
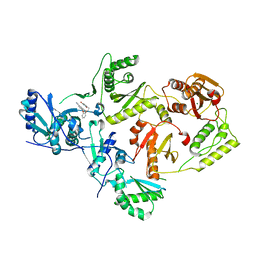 | | Crystal Structure of HIV-1 Reverse Transcriptase (Y181C) varient in Complex with 5-(2-(2-(2,4-dioxo-3,4-dihydropyrimidin-1(2H)-yl)ethoxy)phenoxy)-2-naphthonitrile (JLJ600), a non-nucleoside inhibitor | | Descriptor: | 5-{2-[2-(2,4-dioxo-3,4-dihydropyrimidin-1(2H)-yl)ethoxy]phenoxy}naphthalene-2-carbonitrile, MAGNESIUM ION, Reverse transcriptase/ribonuclease H, ... | | Authors: | Hollander, K, Frey, K.M, Jorgensen, W.L, Anderson, K.S. | | Deposit date: | 2023-05-11 | | Release date: | 2023-11-29 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.955 Å) | | Cite: | Exploring novel HIV-1 reverse transcriptase inhibitors with drug-resistant mutants: A double mutant surprise.
Protein Sci., 32, 2023
|
|
8T2T
 
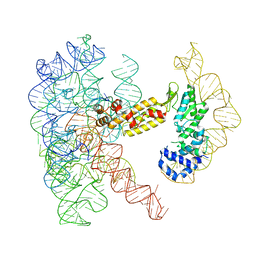 | | Structure of a group II intron ribonucleoprotein in the post-ligation (post-2F) state | | Descriptor: | AMMONIUM ION, Group II intron reverse transcriptase/maturase, MAGNESIUM ION, ... | | Authors: | Xu, L, Liu, T, Chung, K, Pyle, A.M. | | Deposit date: | 2023-06-06 | | Release date: | 2023-11-22 | | Last modified: | 2024-01-03 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Structural insights into intron catalysis and dynamics during splicing.
Nature, 624, 2023
|
|
