4LMZ
 
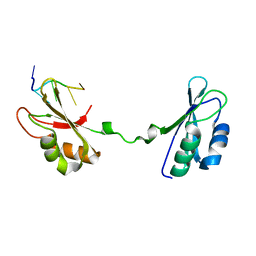 | |
4W90
 
 | | Crystal structure of Bacillus subtilis cyclic-di-AMP riboswitch ydaO | | Descriptor: | (2R,3R,3aS,5R,7aR,9R,10R,10aS,12R,14aR)-2,9-bis(6-amino-9H-purin-9-yl)octahydro-2H,7H-difuro[3,2-d:3',2'-j][1,3,7,9,2,8 ]tetraoxadiphosphacyclododecine-3,5,10,12-tetrol 5,12-dioxide, MAGNESIUM ION, U1 small nuclear ribonucleoprotein A, ... | | Authors: | Jones, C.P, Ferre-D'Amare, A.R. | | Deposit date: | 2014-08-26 | | Release date: | 2014-10-15 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (3.118 Å) | | Cite: | Crystal structure of a c-di-AMP riboswitch reveals an internally pseudo-dimeric RNA.
Embo J., 33, 2014
|
|
4YUD
 
 | |
9GEA
 
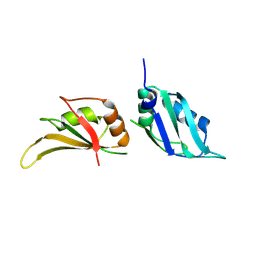 | |
3R1L
 
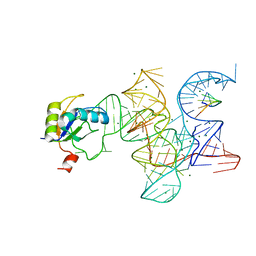 | | Crystal structure of the Class I ligase ribozyme-substrate preligation complex, C47U mutant, Mg2+ bound | | Descriptor: | 5'-R(*UP*CP*CP*AP*GP*UP*A)-3', Class I ligase ribozyme, MAGNESIUM ION, ... | | Authors: | Shechner, D.M, Bartel, D.P. | | Deposit date: | 2011-03-10 | | Release date: | 2011-08-31 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (3.125 Å) | | Cite: | The structural basis of RNA-catalyzed RNA polymerization.
Nat.Struct.Mol.Biol., 18, 2011
|
|
1ZZN
 
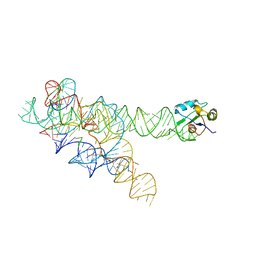 | |
4Y00
 
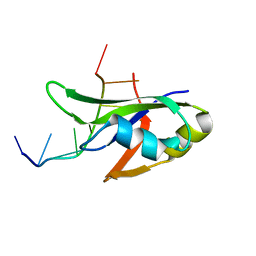 | | Crystal Structure of Human TDP-43 RRM1 Domain with D169G Mutation in Complex with an Unmodified Single-stranded DNA | | Descriptor: | DNA (5'-D(P*TP*TP*GP*AP*GP*CP*GP*T)-3'), TAR DNA-binding protein 43 | | Authors: | Chiang, C.H, Kuo, P.H, Yang, W.Z, Yuan, H.S. | | Deposit date: | 2015-02-05 | | Release date: | 2016-02-10 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structural analysis of disease-related TDP-43 D169G mutation: linking enhanced stability and caspase cleavage efficiency to protein accumulation
Sci Rep, 6, 2016
|
|
3S8S
 
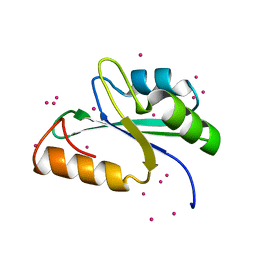 | | Crystal structure of the RRM domain of human SETD1A | | Descriptor: | Histone-lysine N-methyltransferase SETD1A, UNKNOWN ATOM OR ION | | Authors: | Chao, X, Tempel, W, Bian, C, Cerovina, T, Walker, J.R, Bountra, C, Weigelt, J, Arrowsmith, C.H, Edwards, A.M, Min, J, Structural Genomics Consortium (SGC) | | Deposit date: | 2011-05-30 | | Release date: | 2011-06-08 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Crystal structure of the RRM domain of human SETD1A
to be published
|
|
4Y0F
 
 | | Crystal Structure of Human TDP-43 RRM1 Domain in Complex with an Unmodified Single-stranded DNA | | Descriptor: | DNA (5'-D(*GP*TP*TP*GP*AP*GP*CP*GP*TP*T)-3'), TAR DNA-binding protein 43 | | Authors: | Chiang, C.H, Kuo, P.H, Doudeva, L.G, Wang, Y.T, Yuan, H.S. | | Deposit date: | 2015-02-06 | | Release date: | 2016-02-10 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.648 Å) | | Cite: | Structural analysis of disease-related TDP-43 D169G mutation: linking enhanced stability and caspase cleavage efficiency to protein accumulation
Sci Rep, 6, 2016
|
|
4IUF
 
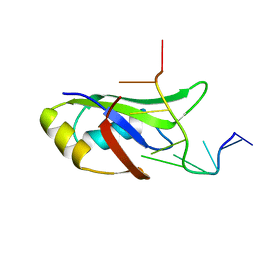 | | Crystal Structure of Human TDP-43 RRM1 Domain in Complex with a Single-stranded DNA | | Descriptor: | 5'-D(*GP*TP*TP*GP*(XUA)P*GP*CP*GP*T)-3', TAR DNA-binding protein 43 | | Authors: | Kuo, P.H, Doudeva, L.G, Wang, Y.T, Yang, W.Z, Yuan, H.S. | | Deposit date: | 2013-01-21 | | Release date: | 2014-01-29 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.752 Å) | | Cite: | The crystal structure of TDP-43 RRM1-DNA complex reveals the specific recognition for UG- and TG-rich nucleic acids.
Nucleic Acids Res., 42, 2014
|
|
4LJM
 
 | |
2X1F
 
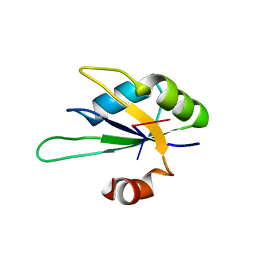 | | Structure of Rna15 RRM with bound RNA (GU) | | Descriptor: | 5'-R(*GP*UP*UP*GP*UP)-3', MRNA 3'-END-PROCESSING PROTEIN RNA15 | | Authors: | Pancevac, C, Goldstone, D.C, Ramos, A, Taylor, I.A. | | Deposit date: | 2010-01-06 | | Release date: | 2010-02-02 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structure of the RNA15 Rrm-RNA Complex Reveals the Molecular Basis of Gu Specificity in Transcriptional 3-End Processing Factors.
Nucleic Acids Res., 38, 2010
|
|
7EB1
 
 | | Solution NMR structure of the RRM domain of RNA binding protein RBM3 from homo sapiens | | Descriptor: | RNA-binding protein 3 | | Authors: | Boral, S, Roy, S, Basak, A.J, Maiti, S, Lee, W, De, S. | | Deposit date: | 2021-03-08 | | Release date: | 2021-12-08 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structural and dynamic studies of the human RNA binding protein RBM3 reveals the molecular basis of its oligomerization and RNA recognition.
Febs J., 289, 2022
|
|
2XS2
 
 | |
1B7F
 
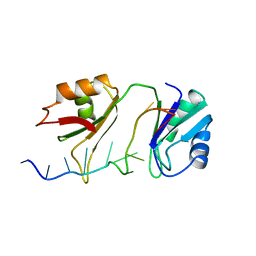 | | SXL-LETHAL PROTEIN/RNA COMPLEX | | Descriptor: | PROTEIN (SXL-LETHAL PROTEIN), RNA (5'-R(P*GP*UP*UP*GP*UP*UP*UP*UP*UP*UP*UP*U)-3') | | Authors: | Handa, N, Nureki, O, Kurimoto, K, Kim, I, Sakamoto, H, Shimura, Y, Muto, Y, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 1999-01-23 | | Release date: | 1999-05-03 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural basis for recognition of the tra mRNA precursor by the Sex-lethal protein.
Nature, 398, 1999
|
|
1AUD
 
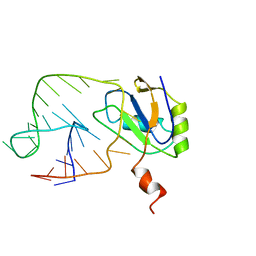 | | U1A-UTRRNA, NMR, 31 STRUCTURES | | Descriptor: | RNA 3UTR, U1A 102 | | Authors: | Allain, F.H.-T, Gubser, C.C, Howe, P.W.A, Nagai, K, Neuhaus, D, Varani, G. | | Deposit date: | 1997-08-22 | | Release date: | 1998-02-25 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Structural basis of the RNA-binding specificity of human U1A protein.
EMBO J., 16, 1997
|
|
7JLY
 
 | |
5WWE
 
 | | Crystal structure of hnRNPA2B1 in complex with RNA | | Descriptor: | Heterogeneous nuclear ribonucleoproteins A2/B1, RNA (5'-R(*AP*GP*GP*GP*AP*CP*UP*AP*GP*A)-3') | | Authors: | Wu, B.X, Su, S.C, Ma, J.B. | | Deposit date: | 2017-01-01 | | Release date: | 2018-01-17 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Molecular basis for the specific and multivariant recognitions of RNA substrates by human hnRNP A2/B1.
Nat Commun, 9, 2018
|
|
5WWG
 
 | | Crystal structure of hnRNPA2B1 in complex with AAGGACUUGC | | Descriptor: | Heterogeneous nuclear ribonucleoproteins A2/B1, RNA (5'-R(*AP*AP*GP*GP*AP*CP*UP*U)-3') | | Authors: | Wu, B.X, Su, S.C, Ma, J.B. | | Deposit date: | 2017-01-01 | | Release date: | 2018-01-17 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | Molecular basis for the specific and multivariant recognitions of RNA substrates by human hnRNP A2/B1.
Nat Commun, 9, 2018
|
|
5MPG
 
 | |
5X3Z
 
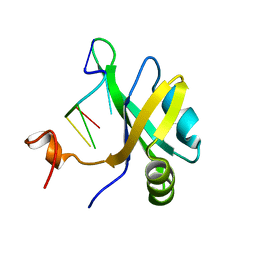 | | Solution structure of musashi1 RBD2 in complex with RNA | | Descriptor: | RNA (5'-R(*GP*UP*AP*GP*U)-3'), RNA-binding protein Musashi homolog 1 | | Authors: | Iwaoka, R, Nagata, T, Tsuda, K, Imai, T, Okano, H, Kobayashi, N, Katahira, M. | | Deposit date: | 2017-02-09 | | Release date: | 2017-12-13 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Structural Insight into the Recognition of r(UAG) by Musashi-1 RBD2, and Construction of a Model of Musashi-1 RBD1-2 Bound to the Minimum Target RNA
Molecules, 22, 2017
|
|
1JMT
 
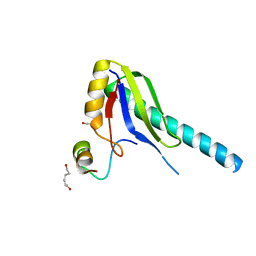 | | X-ray Structure of a Core U2AF65/U2AF35 Heterodimer | | Descriptor: | HEXANE-1,6-DIOL, SPLICING FACTOR U2AF 35 KDA SUBUNIT, SPLICING FACTOR U2AF 65 KDA SUBUNIT | | Authors: | Kielkopf, C.L, Rodionova, N.A, Green, M.R, Burley, S.K. | | Deposit date: | 2001-07-19 | | Release date: | 2001-09-19 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | A novel peptide recognition mode revealed by the X-ray structure of a core U2AF35/U2AF65 heterodimer.
Cell(Cambridge,Mass.), 106, 2001
|
|
1IQT
 
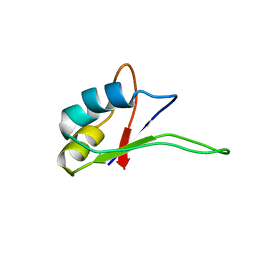 | | Solution structure of the C-terminal RNA-binding domain of heterogeneous nuclear ribonucleoprotein D0 (AUF1) | | Descriptor: | heterogeneous nuclear ribonucleoprotein D0 | | Authors: | Katahira, M, Miyanoiri, Y, Enokizono, Y, Matsuda, G, Nagata, T, Ishikawa, F, Uesugi, S. | | Deposit date: | 2001-08-01 | | Release date: | 2002-08-07 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | Structure of the C-terminal RNA-binding domain of hnRNP D0 (AUF1), its interactions with RNA and DNA, and change in backbone dynamics upon complex formation with DNA.
J.Mol.Biol., 311, 2001
|
|
1L3K
 
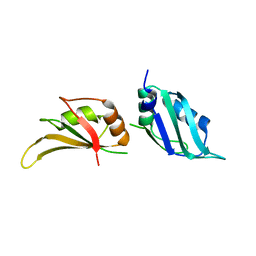 | | UP1, THE TWO RNA-RECOGNITION MOTIF DOMAIN OF HNRNP A1 | | Descriptor: | HETEROGENEOUS NUCLEAR RIBONUCLEOPROTEIN A1 | | Authors: | Vitali, J, Ding, J, Jiang, J, Zhang, Y, Krainer, A.R, Xu, R.-M. | | Deposit date: | 2002-02-27 | | Release date: | 2002-04-17 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | Correlated alternative side chain conformations in the RNA-recognition motif of heterogeneous nuclear ribonucleoprotein A1.
Nucleic Acids Res., 30, 2002
|
|
7S3B
 
 | |
