7XJR
 
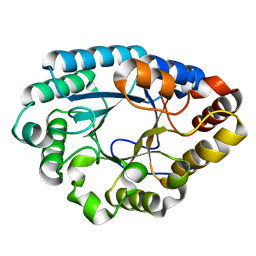 | | MLXase AlXyn26A | | Descriptor: | AlXyn26A | | Authors: | Zhang, Y.Z, Chen, X.L, Zhao, F, Yu, C.M. | | Deposit date: | 2022-04-18 | | Release date: | 2023-04-26 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | A novel class of xylanases specifically degrade marine red algal beta 1,3/1,4-mixed-linkage xylan.
J.Biol.Chem., 299, 2023
|
|
1MLX
 
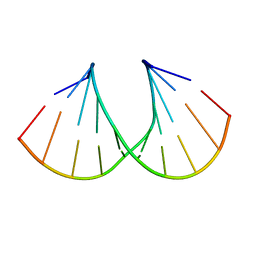 | | Crystal Structure Analysis of a 2'-O-[2-(Methylthio)-ethyl]-Modified Oligodeoxynucleotide Duplex | | Descriptor: | 5'-D(*GP*CP*GP*TP*AP*SMTP*AP*CP*GP*C)-3' | | Authors: | Prakash, T.P, Manoharan, M, Kawasaki, A.M, Fraser, A.S, Lesnik, E.A, Sioufi, N, Leeds, J.M, Teplova, M, Egli, M. | | Deposit date: | 2002-08-31 | | Release date: | 2002-12-04 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | 2'-O-[2-(Methylthio)ethyl]-Modified Oligonucleotide: An Analogue of 2'-O-[2-(Methoxy)-ethyl]-Modified
Oligonucleotide with Improved Protein Binding Properties and High Binding Affinity to Target RNA
Biochemistry, 41, 2002
|
|
6MLX
 
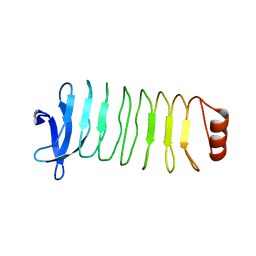 | | Crystal structure of T. pallidum Leucine Rich Repeat protein (TpLRR) | | Descriptor: | Leucine-rich repeat protein TpLRR | | Authors: | Ramaswamy, R, Loveless, B.C, Houston, S, Cameron, C.E, Boulanger, M.J. | | Deposit date: | 2018-09-28 | | Release date: | 2019-07-17 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural characterization of Treponema pallidum Tp0225 reveals an unexpected leucine-rich repeat architecture.
Acta Crystallogr.,Sect.F, 75, 2019
|
|
7MLX
 
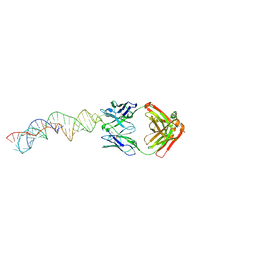 | |
5MLX
 
 | | Open loop conformation of PhaZ7 Y105E mutant | | Descriptor: | CHLORIDE ION, PHB depolymerase PhaZ7, SODIUM ION | | Authors: | Kellici, T, Mavromoustakos, T, Jendrossek, D, Papageorgiou, A.C. | | Deposit date: | 2016-12-08 | | Release date: | 2017-05-10 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystal structure analysis, covalent docking, and molecular dynamics calculations reveal a conformational switch in PhaZ7 PHB depolymerase.
Proteins, 85, 2017
|
|
4MLX
 
 | |
2MLX
 
 | | NMR structure of E. coli Trigger Factor in complex with unfolded PhoA220-310 | | Descriptor: | Alkaline phosphatase, Trigger factor | | Authors: | Saio, T, Guan, X, Rossi, P, Economou, A, Kalodimos, C.G. | | Deposit date: | 2014-03-05 | | Release date: | 2014-05-21 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Structural basis for protein antiaggregation activity of the trigger factor chaperone.
Science, 344, 2014
|
|
3MLX
 
 | | Crystal structure of anti-HIV-1 V3 Fab 3074 in complex with an MN V3 peptide | | Descriptor: | HIV-1 gp120 third variable region (V3) crown, Human monoclonal anti-HIV-1 gp120 V3 antibody 3074 Fab heavy chain, Human monoclonal anti-HIV-1 gp120 V3 antibody 3074 Fab light chain | | Authors: | Kong, X.-P. | | Deposit date: | 2010-04-18 | | Release date: | 2010-07-14 | | Last modified: | 2017-11-08 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Conserved structural elements in the V3 crown of HIV-1 gp120.
Nat.Struct.Mol.Biol., 17, 2010
|
|
4MLT
 
 | |
2MLZ
 
 | | NMR structure of E. coli Trigger Factor in complex with unfolded PhoA365-471 | | Descriptor: | Alkaline phosphatase, Trigger factor | | Authors: | Saio, T, Guan, X, Rossi, P, Economou, A, Kalodimos, C.G. | | Deposit date: | 2014-03-05 | | Release date: | 2014-05-21 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Structural basis for protein antiaggregation activity of the trigger factor chaperone.
Science, 344, 2014
|
|
2MLY
 
 | | NMR structure of E. coli Trigger Factor in complex with unfolded PhoA1-150 | | Descriptor: | Alkaline phosphatase, Trigger factor | | Authors: | Saio, T, Guan, X, Rossi, P, Economou, A, Kalodimos, C.G. | | Deposit date: | 2014-03-05 | | Release date: | 2014-05-21 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Structural basis for protein antiaggregation activity of the trigger factor chaperone.
Science, 344, 2014
|
|
7XS3
 
 | | AlXyn26A E243A-X3X4X | | Descriptor: | AlXyn26A E243A-X3X4X, beta-D-xylopyranose-(1-3)-beta-D-xylopyranose-(1-4)-beta-D-xylopyranose | | Authors: | Zhang, Y.Z, Chen, X.L, Zhao, F, Yu, C.M. | | Deposit date: | 2022-05-12 | | Release date: | 2023-05-17 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | A novel class of xylanases specifically degrade marine red algal beta 1,3/1,4-mixed-linkage xylan.
J.Biol.Chem., 299, 2023
|
|
5YVO
 
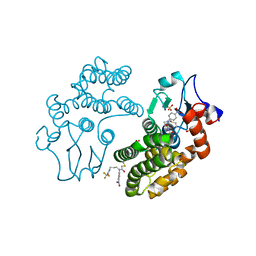 | | Human Glutathione Transferase Omega1 covalently bound to ML175 inhibitor | | Descriptor: | ACETATE ION, Glutathione S-transferase omega-1, N-{3-[(2-chloro-acetyl)-(4-nitro-phenyl)-amino]-propyl}-2,2,2-trifluoro-acetamide, ... | | Authors: | Saisawang, C, Ketterman, A, Wongsantichon, J. | | Deposit date: | 2017-11-27 | | Release date: | 2018-11-28 | | Last modified: | 2022-04-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Glutathione transferase Omega 1-1 (GSTO1-1) modulates Akt and MEK1/2 signaling in human neuroblastoma cell SH-SY5Y.
Proteins, 87, 2019
|
|
5F74
 
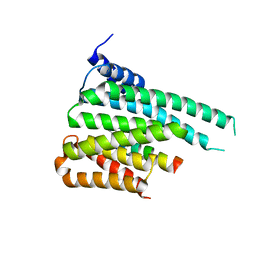 | | Crystal structure of ChREBP:14-3-3 complex bound with AMP | | Descriptor: | 14-3-3 protein beta/alpha, ADENOSINE MONOPHOSPHATE, Carbohydrate-responsive element-binding protein | | Authors: | Jung, H, Uyeda, K. | | Deposit date: | 2015-12-07 | | Release date: | 2016-03-23 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Metabolite Regulation of Nuclear Localization of Carbohydrate-response Element-binding Protein (ChREBP): ROLE OF AMP AS AN ALLOSTERIC INHIBITOR.
J.Biol.Chem., 291, 2016
|
|
6YGJ
 
 | |
4GNT
 
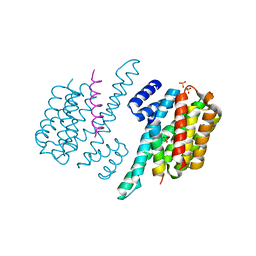 | | Complex of ChREBP and 14-3-3beta | | Descriptor: | 14-3-3 protein beta/alpha, Carbohydrate-responsive element-binding protein, SULFATE ION | | Authors: | Zhang, H, Huang, N. | | Deposit date: | 2012-08-17 | | Release date: | 2012-10-24 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.41 Å) | | Cite: | Crystal Structure of Carbohydrate Response Element Binding Protein (ChREBP) in complex with 14-3-3b
To be Published
|
|
8BTQ
 
 | |
5V3Q
 
 | | Human GSTO1-1 complexed with ML175 | | Descriptor: | Glutathione S-transferase omega-1, N-{3-[(2-chloro-acetyl)-(4-nitro-phenyl)-amino]-propyl}-2,2,2-trifluoro-acetamide, SULFATE ION | | Authors: | Oakley, A.J. | | Deposit date: | 2017-03-07 | | Release date: | 2017-12-20 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | GSTO1-1 plays a pro-inflammatory role in models of inflammation, colitis and obesity.
Sci Rep, 7, 2017
|
|
3FC6
 
 | | hRXRalpha & mLXRalpha with an indole Pharmacophore, SB786875 | | Descriptor: | Nr1h3 protein, RETINOIC ACID, Retinoic acid receptor RXR-alpha, ... | | Authors: | Washburn, D.G, Hoang, T.H, Campobasso, N, Smallwood, A, Parks, D.J, Webb, C.L, Frank, K, Nord, M, Duraiswami, C, Evans, C, Jaye, M, Thompson, S.K. | | Deposit date: | 2008-11-21 | | Release date: | 2009-02-10 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.063 Å) | | Cite: | Synthesis and SAR of potent LXR agonists containing an indole pharmacophore.
Bioorg.Med.Chem.Lett., 19, 2009
|
|
3MLS
 
 | |
3MLZ
 
 | | Crystal structure of anti-HIV-1 V3 Fab 3074 in complex with a VI191 V3 peptide | | Descriptor: | HIV-1 gp120 third variable region (V3) crown, Human monoclonal anti-HIV-1 gp120 V3 antibody 3074 Fab heavy chain, Human monoclonal anti-HIV-1 gp120 V3 antibody 3074 Fab light chain | | Authors: | Kong, X.-P. | | Deposit date: | 2010-04-18 | | Release date: | 2010-07-14 | | Last modified: | 2017-11-08 | | Method: | X-RAY DIFFRACTION (2.99 Å) | | Cite: | Conserved structural elements in the V3 crown of HIV-1 gp120.
Nat.Struct.Mol.Biol., 17, 2010
|
|
3MLW
 
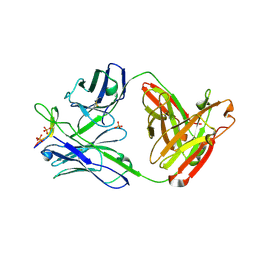 | | Crystal structure of anti-HIV-1 V3 Fab 1006-15D in complex with an MN V3 peptide | | Descriptor: | HIV-1 gp120 third variable region (V3) crown, Human monoclonal anti-HIV-1 gp120 V3 antibody 1006-15D Fab heavy chain, Human monoclonal anti-HIV-1 gp120 V3 antibody 1006-15D Fab light chain, ... | | Authors: | Kong, X.-P. | | Deposit date: | 2010-04-18 | | Release date: | 2010-07-14 | | Last modified: | 2017-11-08 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Conserved structural elements in the V3 crown of HIV-1 gp120.
Nat.Struct.Mol.Biol., 17, 2010
|
|
3MLR
 
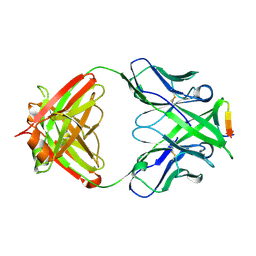 | | Crystal structure of anti-HIV-1 V3 Fab 2557 in complex with a NY5 V3 peptide | | Descriptor: | HIV-1 gp120 third variable region (V3) crown, Human monoclonal anti-HIV-1 gp120 V3 antibody 2557 Fab heavy chain, Human monoclonal anti-HIV-1 gp120 V3 antibody 2557 Fab light chain | | Authors: | Kong, X.-P. | | Deposit date: | 2010-04-18 | | Release date: | 2010-07-14 | | Last modified: | 2017-11-08 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Conserved structural elements in the V3 crown of HIV-1 gp120.
Nat.Struct.Mol.Biol., 17, 2010
|
|
3MLU
 
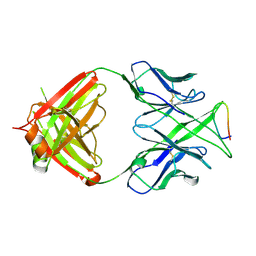 | | Crystal structure of anti-HIV-1 V3 Fab 2557 in complex with a ZAM18 V3 peptide | | Descriptor: | HIV-1 gp120 third variable region (V3) crown, Human monoclonal anti-HIV-1 gp120 V3 antibody 2557 Fab heavy chain, Human monoclonal anti-HIV-1 gp120 V3 antibody 2557 Fab light chain | | Authors: | Kong, X.-P. | | Deposit date: | 2010-04-18 | | Release date: | 2010-07-14 | | Last modified: | 2019-07-17 | | Method: | X-RAY DIFFRACTION (2.77 Å) | | Cite: | Conserved structural elements in the V3 crown of HIV-1 gp120.
Nat.Struct.Mol.Biol., 17, 2010
|
|
3MLV
 
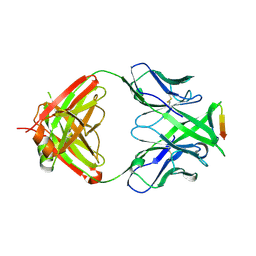 | | Crystal structure of anti-HIV-1 V3 Fab 2557 in complex with an NOF V3 peptide | | Descriptor: | HIV-1 gp120 third variable region (V3) crown, Human monoclonal anti-HIV-1 gp120 V3 antibody 2557 Fab heavy chain, Human monoclonal anti-HIV-1 gp120 V3 antibody 2557 Fab light chain | | Authors: | Kong, X.-P. | | Deposit date: | 2010-04-18 | | Release date: | 2010-07-14 | | Last modified: | 2019-07-17 | | Method: | X-RAY DIFFRACTION (2.48 Å) | | Cite: | Conserved structural elements in the V3 crown of HIV-1 gp120.
Nat.Struct.Mol.Biol., 17, 2010
|
|
