[English] 日本語
 Yorodumi
Yorodumi- EMDB-34174: Structure of beta-arrestin2 in complex with a phosphopeptide corr... -
+ Open data
Open data
- Basic information
Basic information
| Entry | 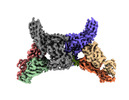 | |||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
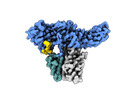
| Title | Structure of beta-arrestin2 in complex with a phosphopeptide corresponding to the human Atypical chemokine receptor 2, ACKR2 (D6R) | |||||||||||||||
 Map data Map data | Full map with pixel size 1.5375 | |||||||||||||||
 Sample Sample |
| |||||||||||||||
 Keywords Keywords |  GPCR / GPCR /  Arrestin / Arrestin /  SIGNALING PROTEIN / SIGNALING PROTEIN /  SIGNALING PROTEIN-IMMUNE SYSTEM complex SIGNALING PROTEIN-IMMUNE SYSTEM complex | |||||||||||||||
| Function / homology |  Function and homology information Function and homology information angiotensin receptor binding / angiotensin receptor binding /  chemokine receptor activity / desensitization of G protein-coupled receptor signaling pathway / C-C chemokine receptor activity / chemokine receptor activity / desensitization of G protein-coupled receptor signaling pathway / C-C chemokine receptor activity /  inositol hexakisphosphate binding / C-C chemokine binding / scavenger receptor activity / G protein-coupled receptor internalization / Chemokine receptors bind chemokines / phosphatidylinositol-3,4,5-trisphosphate binding ... inositol hexakisphosphate binding / C-C chemokine binding / scavenger receptor activity / G protein-coupled receptor internalization / Chemokine receptors bind chemokines / phosphatidylinositol-3,4,5-trisphosphate binding ... angiotensin receptor binding / angiotensin receptor binding /  chemokine receptor activity / desensitization of G protein-coupled receptor signaling pathway / C-C chemokine receptor activity / chemokine receptor activity / desensitization of G protein-coupled receptor signaling pathway / C-C chemokine receptor activity /  inositol hexakisphosphate binding / C-C chemokine binding / scavenger receptor activity / G protein-coupled receptor internalization / Chemokine receptors bind chemokines / phosphatidylinositol-3,4,5-trisphosphate binding / positive regulation of receptor internalization / endocytic vesicle / inositol hexakisphosphate binding / C-C chemokine binding / scavenger receptor activity / G protein-coupled receptor internalization / Chemokine receptors bind chemokines / phosphatidylinositol-3,4,5-trisphosphate binding / positive regulation of receptor internalization / endocytic vesicle /  clathrin-coated pit / clathrin-coated pit /  phosphatidylinositol binding / cell chemotaxis / phosphatidylinositol binding / cell chemotaxis /  actin filament / calcium-mediated signaling / actin filament / calcium-mediated signaling /  receptor internalization / recycling endosome / receptor internalization / recycling endosome /  protein transport / positive regulation of cytosolic calcium ion concentration / protein transport / positive regulation of cytosolic calcium ion concentration /  nuclear membrane / positive regulation of ERK1 and ERK2 cascade / molecular adaptor activity / nuclear membrane / positive regulation of ERK1 and ERK2 cascade / molecular adaptor activity /  early endosome / intracellular signal transduction / early endosome / intracellular signal transduction /  immune response / immune response /  inflammatory response / external side of plasma membrane / intracellular membrane-bounded organelle / inflammatory response / external side of plasma membrane / intracellular membrane-bounded organelle /  signal transduction / signal transduction /  nucleoplasm / nucleoplasm /  nucleus / nucleus /  plasma membrane / plasma membrane /  cytosol / cytosol /  cytoplasm cytoplasmSimilarity search - Function | |||||||||||||||
| Biological species |   Bos taurus (cattle) / Bos taurus (cattle) /   Mus musculus (house mouse) / Mus musculus (house mouse) /   Homo sapiens (human) Homo sapiens (human) | |||||||||||||||
| Method |  single particle reconstruction / single particle reconstruction /  cryo EM / Resolution: 3.35 Å cryo EM / Resolution: 3.35 Å | |||||||||||||||
 Authors Authors | Maharana J / Sarma P / Yadav MK / Banerjee R / Shukla AK | |||||||||||||||
| Funding support |  India, 4 items India, 4 items
| |||||||||||||||
 Citation Citation |  Journal: Science / Year: 2024 Journal: Science / Year: 2024Title: Molecular insights into atypical modes of β-arrestin interaction with seven transmembrane receptors. Authors: Jagannath Maharana / Fumiya K Sano / Parishmita Sarma / Manish K Yadav / Longhan Duan / Tomasz M Stepniewski / Madhu Chaturvedi / Ashutosh Ranjan / Vinay Singh / Sayantan Saha / Gargi ...Authors: Jagannath Maharana / Fumiya K Sano / Parishmita Sarma / Manish K Yadav / Longhan Duan / Tomasz M Stepniewski / Madhu Chaturvedi / Ashutosh Ranjan / Vinay Singh / Sayantan Saha / Gargi Mahajan / Mohamed Chami / Wataru Shihoya / Jana Selent / Ka Young Chung / Ramanuj Banerjee / Osamu Nureki / Arun K Shukla /      Abstract: β-arrestins (βarrs) are multifunctional proteins involved in signaling and regulation of seven transmembrane receptors (7TMRs), and their interaction is driven primarily by agonist-induced receptor ...β-arrestins (βarrs) are multifunctional proteins involved in signaling and regulation of seven transmembrane receptors (7TMRs), and their interaction is driven primarily by agonist-induced receptor activation and phosphorylation. Here, we present seven cryo-electron microscopy structures of βarrs either in the basal state, activated by the muscarinic receptor subtype 2 (M2R) through its third intracellular loop, or activated by the βarr-biased decoy D6 receptor (D6R). Combined with biochemical, cellular, and biophysical experiments, these structural snapshots allow the visualization of atypical engagement of βarrs with 7TMRs and also reveal a structural transition in the carboxyl terminus of βarr2 from a β strand to an α helix upon activation by D6R. Our study provides previously unanticipated molecular insights into the structural and functional diversity encoded in 7TMR-βarr complexes with direct implications for exploring novel therapeutic avenues. | |||||||||||||||
| History |
|
- Structure visualization
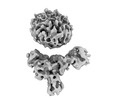
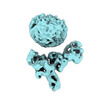
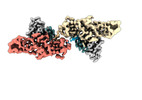
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_34174.map.gz emd_34174.map.gz | 59.5 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-34174-v30.xml emd-34174-v30.xml emd-34174.xml emd-34174.xml | 23.7 KB 23.7 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_34174.png emd_34174.png | 79.5 KB | ||
| Filedesc metadata |  emd-34174.cif.gz emd-34174.cif.gz | 7.2 KB | ||
| Others |  emd_34174_half_map_1.map.gz emd_34174_half_map_1.map.gz emd_34174_half_map_2.map.gz emd_34174_half_map_2.map.gz | 59.2 MB 59.2 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-34174 http://ftp.pdbj.org/pub/emdb/structures/EMD-34174 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-34174 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-34174 | HTTPS FTP |
-Related structure data
| Related structure data |  8go9MC  8j8rC  8j8vC  8j8zC  8j97C  8j9kC  8ja3C  8jafC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_34174.map.gz / Format: CCP4 / Size: 64 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_34174.map.gz / Format: CCP4 / Size: 64 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Full map with pixel size 1.5375 | ||||||||||||||||||||
| Voxel size | X=Y=Z: 1.5375 Å | ||||||||||||||||||||
| Density |
| ||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Half map: Half map A with pixel size 1.5375
| File | emd_34174_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Half map A with pixel size 1.5375 | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: Half map B with pixel size 1.5375
| File | emd_34174_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Half map B with pixel size 1.5375 | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : Peptide bound beta-arrestin2 in complex with Fab30
| Entire | Name: Peptide bound beta-arrestin2 in complex with Fab30 |
|---|---|
| Components |
|
-Supramolecule #1: Peptide bound beta-arrestin2 in complex with Fab30
| Supramolecule | Name: Peptide bound beta-arrestin2 in complex with Fab30 / type: complex / ID: 1 / Parent: 0 / Macromolecule list: all |
|---|---|
| Molecular weight | Theoretical: 190 KDa |
-Supramolecule #2: beta-arrestin2
| Supramolecule | Name: beta-arrestin2 / type: complex / ID: 2 / Parent: 1 / Macromolecule list: #1 |
|---|---|
| Source (natural) | Organism:   Bos taurus (cattle) Bos taurus (cattle) |
-Supramolecule #3: Fab30
| Supramolecule | Name: Fab30 / type: complex / ID: 3 / Parent: 2 / Macromolecule list: #2-#3 |
|---|---|
| Source (natural) | Organism:   Mus musculus (house mouse) Mus musculus (house mouse) |
-Supramolecule #4: Atypical chemokine receptor 2
| Supramolecule | Name: Atypical chemokine receptor 2 / type: complex / ID: 4 / Parent: 3 / Macromolecule list: #4 |
|---|---|
| Source (natural) | Organism:   Homo sapiens (human) / Synthetically produced: Yes Homo sapiens (human) / Synthetically produced: Yes |
-Macromolecule #1: Beta-arrestin-2
| Macromolecule | Name: Beta-arrestin-2 / type: protein_or_peptide / ID: 1 / Number of copies: 2 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:   Bos taurus (cattle) Bos taurus (cattle) |
| Molecular weight | Theoretical: 47.217676 KDa |
| Recombinant expression | Organism:   Escherichia coli (E. coli) Escherichia coli (E. coli) |
| Sequence | String: MGEKPGTRVF KKSSPNGKLT VYLGKRDFVD HLDKVDPVDG VVLVDPDYLK DRKVFVTLTV AFRYGREDCD VLGLSFRKDL FIANYQAFP PTPNPPRPPT RLQERLLRKL GQHAHPFFFT IPQNLPSSVT LQPGPEDTGK ALGVDFEIRA FVAKSLEEKS H KRNSVRLV ...String: MGEKPGTRVF KKSSPNGKLT VYLGKRDFVD HLDKVDPVDG VVLVDPDYLK DRKVFVTLTV AFRYGREDCD VLGLSFRKDL FIANYQAFP PTPNPPRPPT RLQERLLRKL GQHAHPFFFT IPQNLPSSVT LQPGPEDTGK ALGVDFEIRA FVAKSLEEKS H KRNSVRLV IRKVQFAPEK PGPQPSAETT RHFLMSDRSL HLEASLDKEL YYHGEPLNVN VHVTNNSTKT VKKIKVSVRQ YA DIVLFST AQYKVPVAQV EQDDQVSPSS TFSKVYTITP FLANNREKRG LALDGKLKHE DTNLASSTIV KEGANKEVLG ILV SYRVKV KLVVSRGGDV SVELPFVLMH PKPHDHIALP RPQSAATHPP TLLPSAVPET DAPVDTNLIE FETNYATDDD IVFE DFARL RLKGLKDEDY DDQFC UniProtKB:  Beta-arrestin-2 Beta-arrestin-2 |
-Macromolecule #2: Fab30 Heavy Chain
| Macromolecule | Name: Fab30 Heavy Chain / type: protein_or_peptide / ID: 2 / Number of copies: 2 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:   Mus musculus (house mouse) Mus musculus (house mouse) |
| Molecular weight | Theoretical: 25.512354 KDa |
| Recombinant expression | Organism:   Escherichia coli (E. coli) Escherichia coli (E. coli) |
| Sequence | String: EISEVQLVES GGGLVQPGGS LRLSCAASGF NVYSSSIHWV RQAPGKGLEW VASISSYYGY TYYADSVKGR FTISADTSKN TAYLQMNSL RAEDTAVYYC ARSRQFWYSG LDYWGQGTLV TVSSASTKGP SVFPLAPSSK STSGGTAALG CLVKDYFPEP V TVSWNSGA ...String: EISEVQLVES GGGLVQPGGS LRLSCAASGF NVYSSSIHWV RQAPGKGLEW VASISSYYGY TYYADSVKGR FTISADTSKN TAYLQMNSL RAEDTAVYYC ARSRQFWYSG LDYWGQGTLV TVSSASTKGP SVFPLAPSSK STSGGTAALG CLVKDYFPEP V TVSWNSGA LTSGVHTFPA VLQSSGLYSL SSVVTVPSSS LGTQTYICNV NHKPSNTKVD KKVEPKSCDK THHHHHHHH |
-Macromolecule #3: Fab30 Light Chain
| Macromolecule | Name: Fab30 Light Chain / type: protein_or_peptide / ID: 3 / Number of copies: 2 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:   Mus musculus (house mouse) Mus musculus (house mouse) |
| Molecular weight | Theoretical: 23.435064 KDa |
| Recombinant expression | Organism:   Escherichia coli (E. coli) Escherichia coli (E. coli) |
| Sequence | String: SDIQMTQSPS SLSASVGDRV TITCRASQSV SSAVAWYQQK PGKAPKLLIY SASSLYSGVP SRFSGSRSGT DFTLTISSLQ PEDFATYYC QQYKYVPVTF GQGTKVEIKR TVAAPSVFIF PPSDSQLKSG TASVVCLLNN FYPREAKVQW KVDNALQSGN S QESVTEQD ...String: SDIQMTQSPS SLSASVGDRV TITCRASQSV SSAVAWYQQK PGKAPKLLIY SASSLYSGVP SRFSGSRSGT DFTLTISSLQ PEDFATYYC QQYKYVPVTF GQGTKVEIKR TVAAPSVFIF PPSDSQLKSG TASVVCLLNN FYPREAKVQW KVDNALQSGN S QESVTEQD SKDSTYSLSS TLTLSKADYE KHKVYACEVT HQGLSSPVTK SFNRGEC |
-Macromolecule #4: Atypical chemokine receptor 2
| Macromolecule | Name: Atypical chemokine receptor 2 / type: protein_or_peptide / ID: 4 / Number of copies: 2 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:   Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 2.352668 KDa |
| Sequence | String: G(TPO)AQA(SEP)L(SEP)(SEP)C (SEP)E(SEP)(SEP)IL(TPO)A UniProtKB: Atypical chemokine receptor 2 |
-Experimental details
-Structure determination
| Method |  cryo EM cryo EM |
|---|---|
 Processing Processing |  single particle reconstruction single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.4 Component:
| |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Grid | Model: Quantifoil R2/1 / Material: COPPER / Mesh: 300 / Pretreatment - Type: GLOW DISCHARGE / Pretreatment - Time: 45 sec. | |||||||||
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 90 % / Chamber temperature: 283.15 K / Instrument: LEICA EM GP / Details: Blotted for 3 seconds before plunging.. |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD Bright-field microscopy / Cs: 2.7 mm / Nominal defocus max: 2.5 µm / Nominal defocus min: 0.5 µm / Nominal magnification: 165000 Bright-field microscopy / Cs: 2.7 mm / Nominal defocus max: 2.5 µm / Nominal defocus min: 0.5 µm / Nominal magnification: 165000 |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER / Cooling holder cryogen: NITROGEN |
| Image recording | Film or detector model: GATAN K2 SUMMIT (4k x 4k) / Detector mode: COUNTING / Number real images: 10028 / Average electron dose: 56.0 e/Å2 |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
- Image processing
Image processing
| Particle selection | Number selected: 1827629 |
|---|---|
| Startup model | Type of model: PDB ENTRY PDB model - PDB ID: |
| Initial angle assignment | Type: MAXIMUM LIKELIHOOD |
| Final 3D classification | Software - Name: cryoSPARC (ver. 3.3.1) |
| Final angle assignment | Type: MAXIMUM LIKELIHOOD |
| Final reconstruction | Applied symmetry - Point group: C2 (2 fold cyclic ) / Resolution.type: BY AUTHOR / Resolution: 3.35 Å / Resolution method: FSC 0.143 CUT-OFF / Software - Name: cryoSPARC (ver. 3.3.1) / Number images used: 94497 ) / Resolution.type: BY AUTHOR / Resolution: 3.35 Å / Resolution method: FSC 0.143 CUT-OFF / Software - Name: cryoSPARC (ver. 3.3.1) / Number images used: 94497 |
 Movie
Movie Controller
Controller













 Z
Z Y
Y X
X


















