6CPB
 
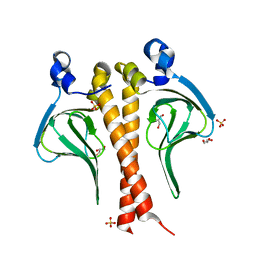 | |
8FG2
 
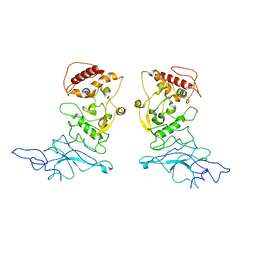 | | SARS-CoV-2 Nucleocapsid dimer structure determined from COVID-19 patients | | 分子名称: | Nucleoprotein | | 著者 | Casasanta, M, Jonaid, G.M, Kaylor, L, Luqiu, W, DiCecco, L, Solares, M, Berry, S, Kelly, D.F. | | 登録日 | 2022-12-12 | | 公開日 | 2023-01-11 | | 最終更新日 | 2023-10-11 | | 実験手法 | ELECTRON MICROSCOPY (6 Å) | | 主引用文献 | Structural Insights of the SARS-CoV-2 Nucleocapsid Protein: Implications for the Inner-workings of Rapid Antigen Tests.
Microsc Microanal, 29, 2023
|
|
8TKL
 
 | | Murine NF-kappaB p50 Rel Homology Region homodimer in complex with a Test 16-mer kappaB-like DNA | | 分子名称: | Nuclear factor NF-kappa-B p50 subunit, Test 17-mer kappaB-like DNA | | 著者 | Mitchel, S, Mealka, M, Rogers, W.E, Milani, C, Acuna, L.M, Huxford, T. | | 登録日 | 2023-07-25 | | 公開日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (3 Å) | | 主引用文献 | X-ray Crystallographic Study of Preferred Spacing by the NF-kappa B p50 Homodimer on kappa B DNA.
Biomolecules, 13, 2023
|
|
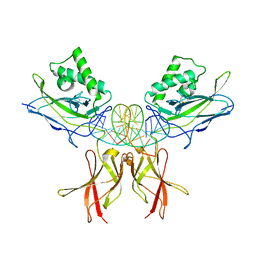
8FD5
 
 | | Nucleocapsid monomer structure from SARS-CoV-2 | | 分子名称: | Nucleoprotein | | 著者 | Casasanta, M, Jonaid, G.M, Kaylor, L, Luqiu, W, DiCecco, L, Solares, M, Berry, S, Kelly, D.F. | | 登録日 | 2022-12-02 | | 公開日 | 2023-01-11 | | 最終更新日 | 2023-10-11 | | 実験手法 | ELECTRON MICROSCOPY (4.57 Å) | | 主引用文献 | Structural Insights of the SARS-CoV-2 Nucleocapsid Protein: Implications for the Inner-workings of Rapid Antigen Tests.
Microsc Microanal, 29, 2023
|
|
1LGW
 
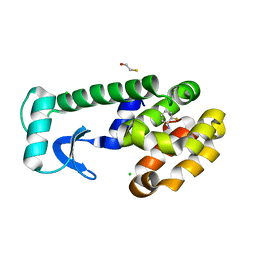 | | T4 Lysozyme Mutant L99A/M102Q Bound by 2-fluoroaniline | | 分子名称: | 2-FLUOROANILINE, BETA-MERCAPTOETHANOL, CHLORIDE ION, ... | | 著者 | Wei, B.Q, Baase, W.A, Weaver, L.H, Matthews, B.W, Shoichet, B.K. | | 登録日 | 2002-04-16 | | 公開日 | 2002-05-08 | | 最終更新日 | 2023-08-16 | | 実験手法 | X-RAY DIFFRACTION (1.85 Å) | | 主引用文献 | A Model Binding Site for Testing Scoring Functions in Molecular Docking
J.Mol.Biol., 322, 2002
|
|
1LGU
 
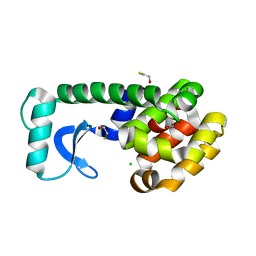 | | T4 Lysozyme Mutant L99A/M102Q | | 分子名称: | BETA-MERCAPTOETHANOL, CHLORIDE ION, Lysozyme | | 著者 | Wei, B.Q, Baase, W.A, Weaver, L.H, Matthews, B.W, Shoichet, B.K. | | 登録日 | 2002-04-16 | | 公開日 | 2002-05-08 | | 最終更新日 | 2024-04-03 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | A Model Binding Site for Testing Scoring Functions in Molecular Docking
J.Mol.Biol., 322, 2002
|
|
1LGX
 
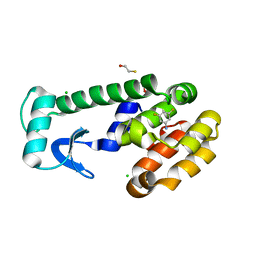 | | T4 Lysozyme Mutant L99A/M102Q Bound by 3,5-difluoroaniline | | 分子名称: | 3,5-DIFLUOROANILINE, BETA-MERCAPTOETHANOL, CHLORIDE ION, ... | | 著者 | Wei, B.Q, Baase, W.A, Weaver, L.H, Matthews, B.W, Shoichet, B.K. | | 登録日 | 2002-04-16 | | 公開日 | 2002-05-08 | | 最終更新日 | 2023-08-16 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | A Model Binding Site for Testing Scoring Functions in Molecular Docking
J.Mol.Biol., 322, 2002
|
|
1LI3
 
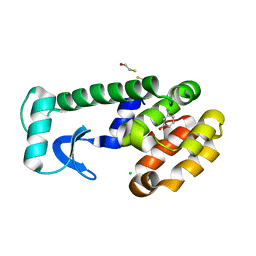 | | T4 lysozyme mutant L99A/M102Q bound by 3-chlorophenol | | 分子名称: | 3-CHLOROPHENOL, BETA-MERCAPTOETHANOL, CHLORIDE ION, ... | | 著者 | Wei, B.Q, Baase, W.A, Weaver, L.H, Matthews, B.W, Shoichet, B.K. | | 登録日 | 2002-04-17 | | 公開日 | 2002-05-08 | | 最終更新日 | 2023-08-16 | | 実験手法 | X-RAY DIFFRACTION (1.85 Å) | | 主引用文献 | A Model Binding Site for Testing Scoring Functions in Molecular Docking
J.Mol.Biol., 322, 2002
|
|
1LI6
 
 | | T4 lysozyme mutant L99A/M102Q bound by 5-methylpyrrole | | 分子名称: | 5-METHYLPYRROLE, BETA-MERCAPTOETHANOL, CHLORIDE ION, ... | | 著者 | Wei, B.Q, Baase, W.A, Weaver, L.H, Matthews, B.W, Shoichet, B.K. | | 登録日 | 2002-04-17 | | 公開日 | 2002-05-08 | | 最終更新日 | 2023-08-16 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | A Model Binding Site for Testing Scoring Functions in Molecular Docking
J.Mol.Biol., 322, 2002
|
|
1LI2
 
 | | T4 Lysozyme Mutant L99A/M102Q Bound by Phenol | | 分子名称: | BETA-MERCAPTOETHANOL, CHLORIDE ION, Lysozyme, ... | | 著者 | Wei, B.Q, Baase, W.A, Weaver, L.H, Matthews, B.W, Shoichet, B.K. | | 登録日 | 2002-04-17 | | 公開日 | 2002-05-08 | | 最終更新日 | 2023-08-16 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | A Model Binding Site for Testing Scoring Functions in Molecular Docking
J.Mol.Biol., 322, 2002
|
|
7U5Z
 
 | | Crystal Structure of HIV-1 Reverse Transcriptase in Complex with JLJ353 | | 分子名称: | 2-chloro-4-({5-[(2,6-difluorophenyl)methyl]-1,3-oxazol-2-yl}amino)benzonitrile, Reverse transcriptase/ribonuclease H, p51 RT | | 著者 | Hollander, K, Carter, Z, Jorgensen, W.L, Anderson, K.S. | | 登録日 | 2022-03-03 | | 公開日 | 2023-03-15 | | 最終更新日 | 2023-10-25 | | 実験手法 | X-RAY DIFFRACTION (2.3 Å) | | 主引用文献 | Design, synthesis, and biological testing of biphenylmethyloxazole inhibitors targeting HIV-1 reverse transcriptase.
Bioorg.Med.Chem.Lett., 84, 2023
|
|
5KP4
 
 | | Crystal Structure of Ketosteroid Isomerase from Pseudomonas putida (pKSI) bound to 19-nortestosterone | | 分子名称: | (8~{R},9~{S},10~{R},13~{S},14~{S},17~{S})-13-methyl-17-oxidanyl-2,6,7,8,9,10,11,12,14,15,16,17-dodecahydro-1~{H}-cyclop enta[a]phenanthren-3-one, Steroid Delta-isomerase | | 著者 | Wu, Y, Boxer, S.G. | | 登録日 | 2016-07-01 | | 公開日 | 2016-09-07 | | 最終更新日 | 2024-03-06 | | 実験手法 | X-RAY DIFFRACTION (1.706 Å) | | 主引用文献 | A Critical Test of the Electrostatic Contribution to Catalysis with Noncanonical Amino Acids in Ketosteroid Isomerase.
J.Am.Chem.Soc., 138, 2016
|
|
5KP1
 
 | |
5KP3
 
 | |
2RB2
 
 | |
2RBV
 
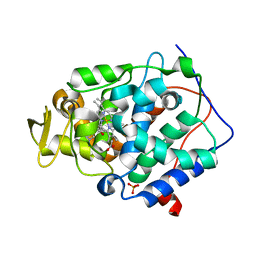 | | Cytochrome C Peroxidase in complex with (1-methyl-1h-pyrrol-2-yl)-methylamine | | 分子名称: | 1-(1-methyl-1H-pyrrol-2-yl)methanamine, Cytochrome C Peroxidase, PHOSPHATE ION, ... | | 著者 | Graves, A.P, Boyce, S.E, Shoichet, B.K. | | 登録日 | 2007-09-19 | | 公開日 | 2008-03-18 | | 最終更新日 | 2023-08-30 | | 実験手法 | X-RAY DIFFRACTION (1.39 Å) | | 主引用文献 | Rescoring docking hit lists for model cavity sites: predictions and experimental testing.
J.Mol.Biol., 377, 2008
|
|
2RBR
 
 | |
2RBS
 
 | |
2RAZ
 
 | |
2RBP
 
 | |
2RC2
 
 | |
2RBY
 
 | | 1-methyl-5-imidazolecarboxaldehyde in complex with Cytochrome C Peroxidase W191G | | 分子名称: | 1-methyl-1H-imidazole-5-carbaldehyde, Cytochrome C Peroxidase, PHOSPHATE ION, ... | | 著者 | Graves, A.P, Boyce, S.E, Shoichet, B.K. | | 登録日 | 2007-09-19 | | 公開日 | 2008-03-18 | | 最終更新日 | 2023-08-30 | | 実験手法 | X-RAY DIFFRACTION (1.5 Å) | | 主引用文献 | Rescoring docking hit lists for model cavity sites: predictions and experimental testing.
J.Mol.Biol., 377, 2008
|
|
2RB0
 
 | | 2,6-difluorobenzylbromide complex with T4 lysozyme L99A | | 分子名称: | 2-(bromomethyl)-1,3-difluorobenzene, Lysozyme, PHOSPHATE ION | | 著者 | Graves, A.P, Boyce, S.E, Shoichet, B.K. | | 登録日 | 2007-09-17 | | 公開日 | 2008-03-18 | | 最終更新日 | 2023-08-30 | | 実験手法 | X-RAY DIFFRACTION (1.84 Å) | | 主引用文献 | Rescoring docking hit lists for model cavity sites: predictions and experimental testing.
J.Mol.Biol., 377, 2008
|
|
2RBQ
 
 | |
2RBO
 
 | |
