2FB2
 
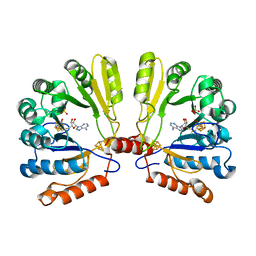 | | Structure of the MoaA Arg17/266/268/Ala triple mutant | | Descriptor: | IRON/SULFUR CLUSTER, Molybdenum cofactor biosynthesis protein A, S-ADENOSYLMETHIONINE, ... | | Authors: | Haenzelmann, P, Schindelin, H. | | Deposit date: | 2005-12-08 | | Release date: | 2006-05-09 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Binding of 5'-GTP to the C-terminal FeS cluster of the radical S-adenosylmethionine enzyme MoaA provides insights into its mechanism
Proc.Natl.Acad.Sci.USA, 103, 2006
|
|
2A5H
 
 | | 2.1 Angstrom X-ray crystal structure of lysine-2,3-aminomutase from Clostridium subterminale SB4, with Michaelis analog (L-alpha-lysine external aldimine form of pyridoxal-5'-phosphate). | | Descriptor: | IRON/SULFUR CLUSTER, L-lysine 2,3-aminomutase, LYSINE, ... | | Authors: | Lepore, B.W, Ruzicka, F.J, Frey, P.A, Ringe, D. | | Deposit date: | 2005-06-30 | | Release date: | 2005-10-04 | | Last modified: | 2017-10-11 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | The X-ray crystal structure of lysine-2,3-aminomutase from Clostridium subterminale.
Proc.Natl.Acad.Sci.Usa, 102, 2005
|
|
1TV8
 
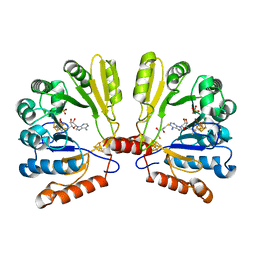 | | Structure of MoaA in complex with S-adenosylmethionine | | Descriptor: | (2R,3S)-1,4-DIMERCAPTOBUTANE-2,3-DIOL, IRON/SULFUR CLUSTER, Molybdenum cofactor biosynthesis protein A, ... | | Authors: | Haenzelmann, P, Schindelin, H. | | Deposit date: | 2004-06-28 | | Release date: | 2004-08-31 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of the S-adenosylmethionine-dependent enzyme MoaA and its implications for molybdenum cofactor deficiency in humans.
Proc.Natl.Acad.Sci.Usa, 101, 2004
|
|
1TV7
 
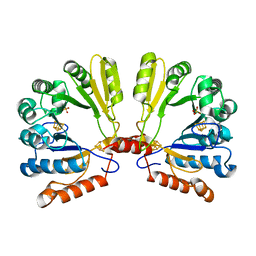 | | Structure of the S-adenosylmethionine dependent Enzyme MoaA | | Descriptor: | IRON/SULFUR CLUSTER, Molybdenum cofactor biosynthesis protein A, SULFATE ION | | Authors: | Haenzelmann, P, Schindelin, H. | | Deposit date: | 2004-06-28 | | Release date: | 2004-08-31 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal structure of the S-adenosylmethionine-dependent enzyme MoaA and its implications for molybdenum cofactor deficiency in humans.
Proc.Natl.Acad.Sci.Usa, 101, 2004
|
|
1R30
 
 | | The Crystal Structure of Biotin Synthase, an S-Adenosylmethionine-Dependent Radical Enzyme | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 6-(5-METHYL-2-OXO-IMIDAZOLIDIN-4-YL)-HEXANOIC ACID, Biotin synthase, ... | | Authors: | Berkovitch, F, Nicolet, Y, Wan, J.T, Jarrett, J.T, Drennan, C.L. | | Deposit date: | 2003-09-30 | | Release date: | 2004-01-13 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | Crystal structure of biotin synthase, an S-adenosylmethionine-dependent radical enzyme.
Science, 303, 2004
|
|
1OLT
 
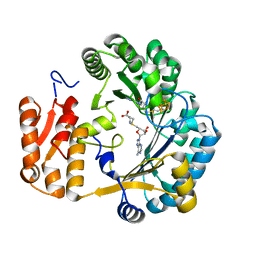 | | Coproporphyrinogen III oxidase (HemN) from Escherichia coli is a Radical SAM enzyme. | | Descriptor: | IRON/SULFUR CLUSTER, OXYGEN-INDEPENDENT COPROPORPHYRINOGEN III OXIDASE, S-ADENOSYLMETHIONINE | | Authors: | Layer, G, Moser, J, Heinz, D.W, Jahn, D, Schubert, W.-D. | | Deposit date: | 2003-08-13 | | Release date: | 2003-12-04 | | Last modified: | 2019-05-08 | | Method: | X-RAY DIFFRACTION (2.07 Å) | | Cite: | Crystal Structure of Coproporphyrinogen III Oxidase Reveals Cofactor Geometry of Radical Sam Enzymes
Embo J., 22, 2003
|
|
