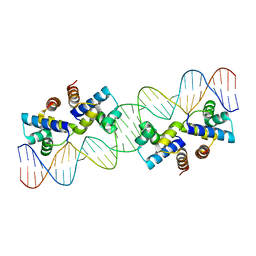
4X4H
 
 | |
2ICT
 
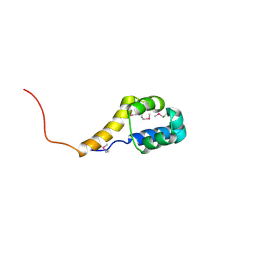 | | Crystal structure of the bacterial antitoxin HigA from Escherichia coli at pH 8.5. Northeast Structural Genomics TARGET ER390. | | Descriptor: | antitoxin higa | | Authors: | Arbing, M.A, Abashidze, M, Hurley, J.M, Zhao, L, Janjua, H, Cunningham, K, Ma, L.C, Xiao, R, Liu, J, Baran, M.C, Acton, T.B, Rost, B, Inouye, M, Woychik, N.A, Montelione, G.T, Hunt, J.F, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2006-09-13 | | Release date: | 2006-09-26 | | Last modified: | 2017-10-18 | | Method: | X-RAY DIFFRACTION (1.63 Å) | | Cite: | Crystal Structures of Phd-Doc, HigA, and YeeU Establish Multiple Evolutionary Links between Microbial Growth-Regulating Toxin-Antitoxin Systems.
Structure, 18, 2010
|
|
4X4D
 
 | |
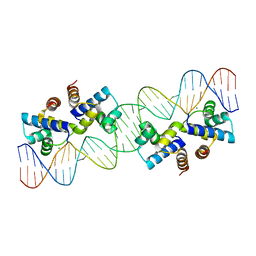
4X4B
 
 | |
4X4C
 
 | |
2JVL
 
 | | NMR structure of the C-terminal domain of MBF1 of Trichoderma reesei | | Descriptor: | TrMBF1 | | Authors: | Kopke Salinas, R, Tomaselli, S, Camilo, C.M, Valencia, E.Y, Farah, C.S, El-Dorry, H, Chambergo, F.S. | | Deposit date: | 2007-09-20 | | Release date: | 2008-09-02 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the C-terminal domain of multiprotein bridging factor 1 (MBF1) of Trichoderma reesei.
Proteins, 75, 2009
|
|
4YBA
 
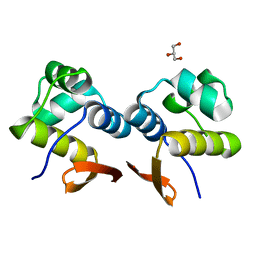 | | The structure of the C.Kpn2I controller protein | | Descriptor: | GLYCEROL, Regulatory protein C | | Authors: | Shevtsov, M.B, Martin, R.N, Swiderska, A, McGeehan, G.E, Kneale, G.G. | | Deposit date: | 2015-02-18 | | Release date: | 2016-03-02 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | The structure of the C.kpn2I controller protein
To Be Published
|
|
4YG1
 
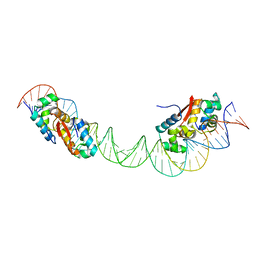 | | HipB-O1-O2 complex/P21212 crystal form | | Descriptor: | Antitoxin HipB, DNA (48-MER) | | Authors: | Schumacher, M.A. | | Deposit date: | 2015-02-25 | | Release date: | 2015-07-29 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (3.25 Å) | | Cite: | HipBA-promoter structures reveal the basis of heritable multidrug tolerance.
Nature, 524, 2015
|
|
2KPJ
 
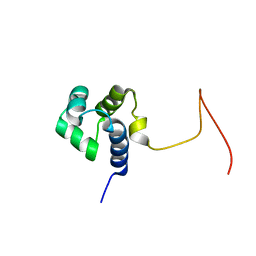 | | Solution Structure Of Protein SOS-response transcriptional repressor, LexA From Eubacterium rectale. Northeast Structural Genomics Consortium Target ErR9A | | Descriptor: | SOS-response transcriptional repressor, LexA | | Authors: | Wu, Y, Eletsky, A, Lee, D, Ghosh, A, Buchwald, W, Zhang, Q, Janjua, H, Garcia, E, Nair, R, Sukumaran, D, Rost, B, Acton, T, Xiao, R, Everett, J, Montelione, G.T, Szyperski, T, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2009-10-16 | | Release date: | 2009-11-17 | | Last modified: | 2024-05-08 | | Method: | SOLUTION NMR | | Cite: | Solution Structure Of Protein SOS-response transcriptional repressor, LexA From Eubacterium rectale. Northeast Structural Genomics Consortium Target ErR9A
To be Published
|
|
4YG4
 
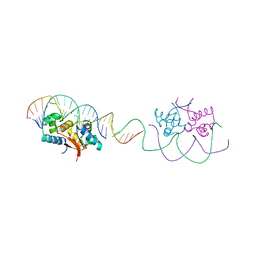 | | HipB-O1-O1* complex | | Descriptor: | Antitoxin HipB, DNA (28-MER), DNA (5'-D(*AP*TP*AP*TP*CP*CP*CP*CP*TP*TP*AP*AP*GP*GP*GP*GP*AP*TP*AP*A)-3') | | Authors: | Schumacher, M.A. | | Deposit date: | 2015-02-25 | | Release date: | 2015-07-29 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | HipBA-promoter structures reveal the basis of heritable multidrug tolerance.
Nature, 524, 2015
|
|
2L49
 
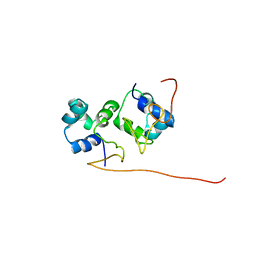 | | The solution structure of the P2 C,the immunity repressor of the P2 bacteriophage | | Descriptor: | C protein | | Authors: | Massad, T, Papadopolos, E, Stenmark, P, Damberg, P. | | Deposit date: | 2010-10-01 | | Release date: | 2010-10-13 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | The C repressor of the P2 bacteriophage.
J.Biomol.Nmr, 64, 2016
|
|
2LYP
 
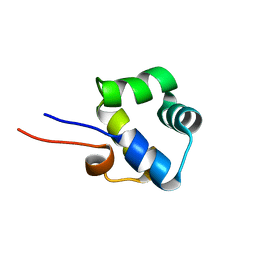 | | NOE-based 3D structure of the monomer of CylR2 in equilibrium with predissociated homodimer at 266K (-7 Celsius degrees) | | Descriptor: | CylR2 | | Authors: | Jaremko, M, Jaremko, L, Kim, H, Cho, M, Schwieters, C.D, Giller, K, Becker, S, Zweckstetter, M. | | Deposit date: | 2012-09-19 | | Release date: | 2013-02-20 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Cold denaturation of a protein dimer monitored at atomic resolution.
Nat.Chem.Biol., 9, 2013
|
|
2LYQ
 
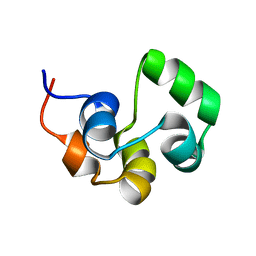 | | NOE-based 3D structure of the monomeric intermediate of CylR2 at 262K (-11 Celsius degrees) | | Descriptor: | CylR2 | | Authors: | Jaremko, M, Jaremko, L, Kim, H, Cho, M, Schwieters, C.D, Giller, K, Becker, S, Zweckstetter, M. | | Deposit date: | 2012-09-19 | | Release date: | 2013-02-20 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Cold denaturation of a protein dimer monitored at atomic resolution.
Nat.Chem.Biol., 9, 2013
|
|
2LYR
 
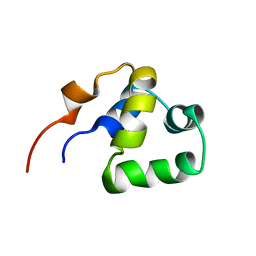 | | NOE-based 3D structure of the monomeric partially-folded intermediate of CylR2 at 259K (-14 Celsius degrees) | | Descriptor: | CylR2 | | Authors: | Jaremko, M, Jaremko, L, Kim, H, Cho, M, Schwieters, C.D, Giller, K, Becker, S, Zweckstetter, M. | | Deposit date: | 2012-09-19 | | Release date: | 2013-02-20 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Cold denaturation of a protein dimer monitored at atomic resolution.
Nat.Chem.Biol., 9, 2013
|
|
2MEZ
 
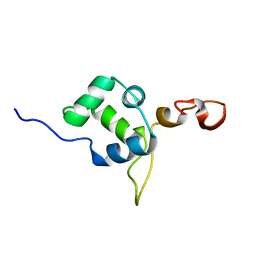 | | Flexible anchoring of archaeal MBF1 on ribosomes suggests role as recruitment factor | | Descriptor: | Multiprotein Bridging Factor (MBP-like) | | Authors: | Launay, H, Blombarch, F, Camilloni, C, Vendruscolo, M, van des Oost, J, Christodoulou, J. | | Deposit date: | 2013-10-03 | | Release date: | 2014-06-25 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Archaeal MBF1 binds to 30S and 70S ribosomes via its helix-turn-helix domain.
Biochem.J., 462, 2014
|
|
2LYK
 
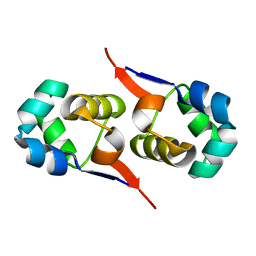 | | NOE-based 3D structure of the CylR2 homodimer at 270K (-3 Celsius degrees) | | Descriptor: | CylR2 | | Authors: | Jaremko, M, Jaremko, L, Kim, H, Cho, M, Schwieters, C.D, Giller, K, Becker, S, Zweckstetter, M. | | Deposit date: | 2012-09-19 | | Release date: | 2013-02-20 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Cold denaturation of a protein dimer monitored at atomic resolution.
Nat.Chem.Biol., 9, 2013
|
|
2LYL
 
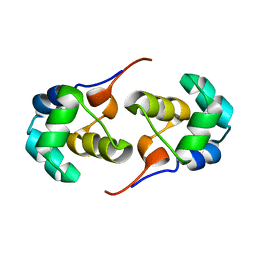 | | NOE-based 3D structure of the predissociated homodimer of CylR2 in equilibrium with monomer at 266K (-7 Celsius degrees) | | Descriptor: | CylR2 | | Authors: | Jaremko, M, Jaremko, L, Kim, H, Cho, M, Schwieters, C.D, Giller, K, Becker, S, Zweckstetter, M. | | Deposit date: | 2012-09-19 | | Release date: | 2013-02-20 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Cold denaturation of a protein dimer monitored at atomic resolution.
Nat.Chem.Biol., 9, 2013
|
|
2LYS
 
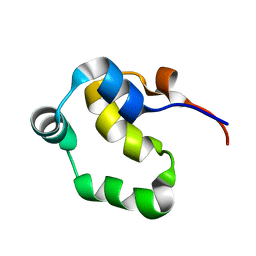 | | NOE-based 3D structure of the monomeric partially-folded intermediate of CylR2 at 257K (-16 Celsius degrees) | | Descriptor: | CylR2 | | Authors: | Jaremko, M, Jaremko, L, Kim, H, Cho, M, Schwieters, C.D, Giller, K, Becker, S, Zweckstetter, M. | | Deposit date: | 2012-09-19 | | Release date: | 2013-02-20 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Cold denaturation of a protein dimer monitored at atomic resolution.
Nat.Chem.Biol., 9, 2013
|
|
2LYJ
 
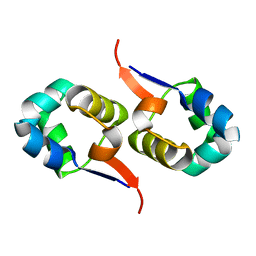 | | NOE-based 3D structure of the CylR2 homodimer at 298K | | Descriptor: | CylR2 | | Authors: | Jaremko, M, Jaremko, L, Kim, H, Cho, M, Giller, K, Becker, S, Zweckstetter, M, Schwieters, C.D. | | Deposit date: | 2012-09-19 | | Release date: | 2013-02-20 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Cold denaturation of a protein dimer monitored at atomic resolution.
Nat.Chem.Biol., 9, 2013
|
|
1X57
 
 | | Solution structures of the HTH domain of human EDF-1 protein | | Descriptor: | Endothelial differentiation-related factor 1 | | Authors: | Nameki, N, Sato, M, Tochio, N, Koshiba, S, Inoue, M, Kigawa, T, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2005-05-15 | | Release date: | 2005-11-15 | | Last modified: | 2022-03-02 | | Method: | SOLUTION NMR | | Cite: | Solution structures of the HTH domain of human EDF-1 protein
To be Published
|
|
1Y7Y
 
 | | High-resolution crystal structure of the restriction-modification controller protein C.AhdI from Aeromonas hydrophila | | Descriptor: | C.AhdI | | Authors: | McGeehan, J.E, Streeter, S.D, Papapanagiotou, I, Fox, G.C, Kneale, G.G. | | Deposit date: | 2004-12-10 | | Release date: | 2005-02-22 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.69 Å) | | Cite: | High-resolution crystal structure of the restriction-modification controller protein C.AhdI from Aeromonas hydrophila.
J.Mol.Biol., 346, 2005
|
|
1ZUG
 
 | | STRUCTURE OF PHAGE 434 CRO PROTEIN, NMR, 20 STRUCTURES | | Descriptor: | PHAGE 434 CRO PROTEIN | | Authors: | Padmanabhan, S, Jimenez, M.A, Gonzalez, C, Sanz, J.M, Gimenez-Gallego, G, Rico, M. | | Deposit date: | 1997-03-14 | | Release date: | 1997-07-07 | | Last modified: | 2022-03-02 | | Method: | SOLUTION NMR | | Cite: | Three-dimensional solution structure and stability of phage 434 Cro protein.
Biochemistry, 36, 1997
|
|
5IFG
 
 | | Crystal structure of HigA-HigB complex from E. Coli | | Descriptor: | Antitoxin HigA, mRNA interferase HigB | | Authors: | Yang, J.S, Zhou, K, Gao, z.Q, Liu, Q.S, Dong, Y.H. | | Deposit date: | 2016-02-26 | | Release date: | 2017-03-01 | | Method: | X-RAY DIFFRACTION (2.702 Å) | | Cite: | Structural insight into the E. coli HigBA complex
Biochem. Biophys. Res. Commun., 478, 2016
|
|
7ZCV
 
 | | Rgg144 of Streptococcus pneumoniae | | Descriptor: | Transcriptional regulator | | Authors: | Wallis, R, Girija, U.V, Yesilkaya, H. | | Deposit date: | 2022-03-29 | | Release date: | 2022-06-01 | | Last modified: | 2022-07-06 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure-function analysis for the development of peptide inhibitors for a Gram-positive quorum sensing system.
Mol.Microbiol., 117, 2022
|
|
1B0N
 
 | | SINR PROTEIN/SINI PROTEIN COMPLEX | | Descriptor: | PROTEIN (SINI PROTEIN), PROTEIN (SINR PROTEIN), ZINC ION | | Authors: | Lewis, R.J, Brannigan, J.A, Offen, W.A, Smith, I, Wilkinson, A.J. | | Deposit date: | 1998-11-11 | | Release date: | 1999-01-13 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | An evolutionary link between sporulation and prophage induction in the structure of a repressor:anti-repressor complex.
J.Mol.Biol., 283, 1998
|
|
