4JRP
 
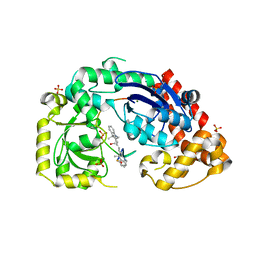 | |
4L8R
 
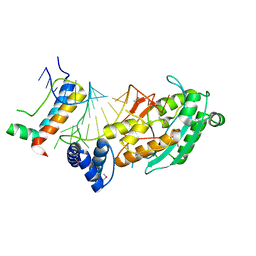 | | Structure of mrna stem-loop, human stem-loop binding protein and 3'hexo ternary complex | | 分子名称: | 3'-5' exoribonuclease 1, HISTONE MRNA STEM-LOOP, Histone RNA hairpin-binding protein | | 著者 | Tan, D, Tong, L. | | 登録日 | 2013-06-17 | | 公開日 | 2013-07-10 | | 最終更新日 | 2017-11-15 | | 実験手法 | X-RAY DIFFRACTION (2.6 Å) | | 主引用文献 | Structure of Histone Mrna Stem-Loop, Human Stem-Loop Binding Protein, and 3'Hexo Ternary Complex.
Science, 339, 2013
|
|
6R9J
 
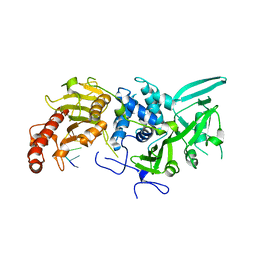 | | Structure of Saccharomyces cerevisiae apo Pan2 pseudoubiquitin hydrolase-RNA exonuclease (UCH-Exo) module in complex with A7 RNA | | 分子名称: | A7 RNA, PAN2-PAN3 deadenylation complex catalytic subunit PAN2 | | 著者 | Tang, T.T.L, Stowell, J.A.W, Hill, C.H, Passmore, L.A. | | 登録日 | 2019-04-03 | | 公開日 | 2019-05-22 | | 最終更新日 | 2024-01-24 | | 実験手法 | X-RAY DIFFRACTION (3.326 Å) | | 主引用文献 | The intrinsic structure of poly(A) RNA determines the specificity of Pan2 and Caf1 deadenylases.
Nat.Struct.Mol.Biol., 26, 2019
|
|
6R9P
 
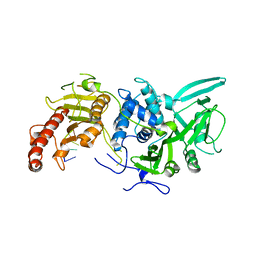 | | Structure of Saccharomyces cerevisiae apo Pan2 pseudoubiquitin hydrolase-RNA exonuclease (UCH-Exo) module in complex with AAUUAA RNA | | 分子名称: | AAUUAA RNA, PAN2-PAN3 deadenylation complex catalytic subunit PAN2 | | 著者 | Tang, T.T.L, Stowell, J.A.W, Hill, C.H, Passmore, L.A. | | 登録日 | 2019-04-03 | | 公開日 | 2019-05-22 | | 最終更新日 | 2024-01-24 | | 実験手法 | X-RAY DIFFRACTION (2.98 Å) | | 主引用文献 | The intrinsic structure of poly(A) RNA determines the specificity of Pan2 and Caf1 deadenylases.
Nat.Struct.Mol.Biol., 26, 2019
|
|
6R9O
 
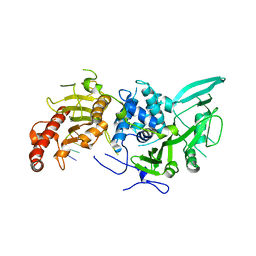 | | Structure of Saccharomyces cerevisiae apo Pan2 pseudoubiquitin hydrolase-RNA exonuclease (UCH-Exo) module in complex with AAGGA RNA | | 分子名称: | AAGGA RNA, PAN2-PAN3 deadenylation complex catalytic subunit PAN2 | | 著者 | Tang, T.T.L, Stowell, J.A.W, Hill, C.H, Passmore, L.A. | | 登録日 | 2019-04-03 | | 公開日 | 2019-05-22 | | 最終更新日 | 2024-01-24 | | 実験手法 | X-RAY DIFFRACTION (3.319 Å) | | 主引用文献 | The intrinsic structure of poly(A) RNA determines the specificity of Pan2 and Caf1 deadenylases.
Nat.Struct.Mol.Biol., 26, 2019
|
|
6R9I
 
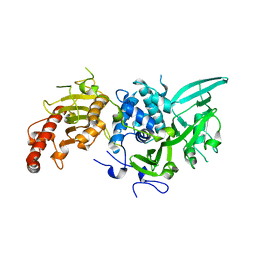 | |
6R9M
 
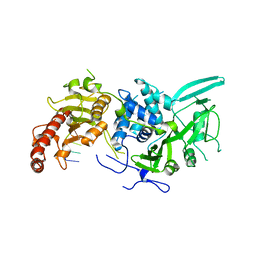 | | Structure of Saccharomyces cerevisiae apo Pan2 pseudoubiquitin hydrolase-RNA exonuclease (UCH-Exo) module in complex with AAGGAA RNA | | 分子名称: | AAGGAA RNA, PAN2-PAN3 deadenylation complex catalytic subunit PAN2 | | 著者 | Tang, T.T.L, Stowell, J.A.W, Hill, C.H, Passmore, L.A. | | 登録日 | 2019-04-03 | | 公開日 | 2019-05-22 | | 最終更新日 | 2024-01-24 | | 実験手法 | X-RAY DIFFRACTION (3.329 Å) | | 主引用文献 | The intrinsic structure of poly(A) RNA determines the specificity of Pan2 and Caf1 deadenylases.
Nat.Struct.Mol.Biol., 26, 2019
|
|
6R9Q
 
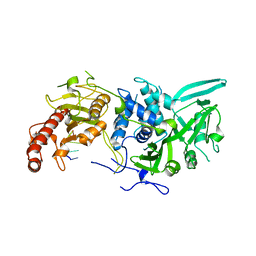 | | Structure of Saccharomyces cerevisiae apo Pan2 pseudoubiquitin hydrolase-RNA exonuclease (UCH-Exo) module in complex with AACCAA RNA | | 分子名称: | AACCAA, PAN2-PAN3 deadenylation complex catalytic subunit PAN2 | | 著者 | Tang, T.T.L, Stowell, J.A.W, Hill, C.H, Passmore, L.A. | | 登録日 | 2019-04-03 | | 公開日 | 2019-05-22 | | 最終更新日 | 2024-01-24 | | 実験手法 | X-RAY DIFFRACTION (3.079 Å) | | 主引用文献 | The intrinsic structure of poly(A) RNA determines the specificity of Pan2 and Caf1 deadenylases.
Nat.Struct.Mol.Biol., 26, 2019
|
|
5M1S
 
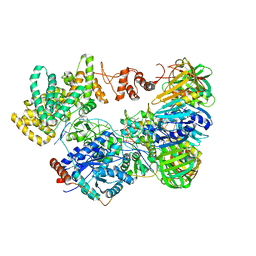 | | Cryo-EM structure of the E. coli replicative DNA polymerase-clamp-exonuclase-theta complex bound to DNA in the editing mode | | 分子名称: | DNA Primer Strand, DNA Template Strand, DNA polymerase III subunit alpha, ... | | 著者 | Fernandez-Leiro, R, Conrad, J, Scheres, S.H.W, Lamers, M.H. | | 登録日 | 2016-10-10 | | 公開日 | 2017-01-18 | | 最終更新日 | 2024-05-15 | | 実験手法 | ELECTRON MICROSCOPY (6.7 Å) | | 主引用文献 | Self-correcting mismatches during high-fidelity DNA replication.
Nat. Struct. Mol. Biol., 24, 2017
|
|
5FKU
 
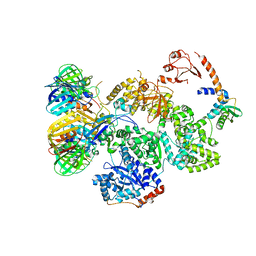 | | cryo-EM structure of the E. coli replicative DNA polymerase complex in DNA free state (DNA polymerase III alpha, beta, epsilon, tau complex) | | 分子名称: | DNA POLYMERASE III SUBUNIT ALPHA, DNA POLYMERASE III SUBUNIT BETA, DNA POLYMERASE III SUBUNIT EPSILON, ... | | 著者 | Fernandez-Leiro, R, Conrad, J, Scheres, S.H.W, Lamers, M.H. | | 登録日 | 2015-10-20 | | 公開日 | 2015-11-25 | | 最終更新日 | 2024-05-08 | | 実験手法 | ELECTRON MICROSCOPY (8.34 Å) | | 主引用文献 | cryo-EM structures of theE. colireplicative DNA polymerase reveal its dynamic interactions with the DNA sliding clamp, exonuclease andtau.
Elife, 4, 2015
|
|
5FKW
 
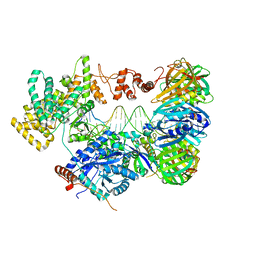 | | cryo-EM structure of the E. coli replicative DNA polymerase complex bound to DNA (DNA polymerase III alpha, beta, epsilon) | | 分子名称: | DNA POLYMERASE III ALPHA, DNA POLYMERASE III BETA, DNA POLYMERASE III EPSILON, ... | | 著者 | Fernandez-Leiro, R, Conrad, J, Scheres, S.H.W, Lamers, M.H. | | 登録日 | 2015-10-20 | | 公開日 | 2015-11-25 | | 最終更新日 | 2024-05-08 | | 実験手法 | ELECTRON MICROSCOPY (7.3 Å) | | 主引用文献 | cryo-EM structures of theE. colireplicative DNA polymerase reveal its dynamic interactions with the DNA sliding clamp, exonuclease andtau.
Elife, 4, 2015
|
|
5FKV
 
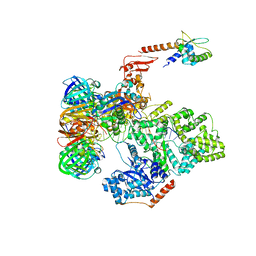 | | cryo-EM structure of the E. coli replicative DNA polymerase complex bound to DNA (DNA polymerase III alpha, beta, epsilon, tau complex) | | 分子名称: | DNA POLYMERASE III BETA, DNA POLYMERASE III EPSILON, DNA POLYMERASE III SUBUNIT ALPHA, ... | | 著者 | Fernandez-Leiro, R, Conrad, J, Scheres, S.H.W, Lamers, M.H. | | 登録日 | 2015-10-20 | | 公開日 | 2015-11-25 | | 最終更新日 | 2024-05-08 | | 実験手法 | ELECTRON MICROSCOPY (8.04 Å) | | 主引用文献 | cryo-EM structures of theE. colireplicative DNA polymerase reveal its dynamic interactions with the DNA sliding clamp, exonuclease andtau.
Elife, 4, 2015
|
|
6R5K
 
 | |
