6SYX
 
 | | Hydrogenase-2 variant R479K - reduced sample exposed to pure oxygen | | Descriptor: | CARBONMONOXIDE-(DICYANO) IRON, CHLORIDE ION, FE3-S4 CLUSTER, ... | | Authors: | Carr, S.B, Beaton, S.E, Evans, R.M, Armstrong, F.A. | | Deposit date: | 2019-10-01 | | Release date: | 2020-10-14 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Hydrogen activation by NiFe-hydrogenases - consolidating the role of the pendant arginine.
To Be Published
|
|
6SYO
 
 | | Hydrogenase-2 variant R479K - As Isolated form | | Descriptor: | CARBONMONOXIDE-(DICYANO) IRON, CHLORIDE ION, FE3-S4 CLUSTER, ... | | Authors: | Carr, S.B, Beaton, S.E, Evans, R.M, Armstrong, F.A. | | Deposit date: | 2019-09-30 | | Release date: | 2020-10-14 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | Hydrogen activation by NiFe-hydrogenases - consolidating the role of the pendant arginine.
To Be Published
|
|
6RUC
 
 | |
6RU9
 
 | |
6RTP
 
 | |
6RP2
 
 | | Threonine to Cysteine (T225C) variant of E coli hydrogenase-1 | | Descriptor: | CARBONMONOXIDE-(DICYANO) IRON, CHLORIDE ION, DODECYL-BETA-D-MALTOSIDE, ... | | Authors: | Carr, S.B, Armstrong, F.A, Zhang, L. | | Deposit date: | 2019-05-13 | | Release date: | 2020-03-25 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Aerobic Photocatalytic H2Production by a [NiFe] Hydrogenase Engineered to Place a Silver Nanocluster in the Electron Relay.
J.Am.Chem.Soc., 142, 2020
|
|
6QII
 
 | | Xenon derivatization of the F420-reducing [NiFe] hydrogenase complex from Methanosarcina barkeri | | Descriptor: | (R,R)-2,3-BUTANEDIOL, Coenzyme F420 hydrogenase subunit alpha, Coenzyme F420 hydrogenase subunit beta, ... | | Authors: | Ilina, Y, Lorent, C, Katz, S, Jeoung, J.H, Shima, S, Horch, M, Zebger, I, Dobbek, H. | | Deposit date: | 2019-01-19 | | Release date: | 2019-10-23 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.28 Å) | | Cite: | X-ray Crystallography and Vibrational Spectroscopy Reveal the Key Determinants of Biocatalytic Dihydrogen Cycling by [NiFe] Hydrogenases.
Angew.Chem.Int.Ed.Engl., 58, 2019
|
|
6QGT
 
 | | The carbon monoxide inhibition of F420-reducing [NiFe] hydrogenase complex from Methanosarcina barkeri | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, (R,R)-2,3-BUTANEDIOL, Coenzyme F420 hydrogenase subunit beta, ... | | Authors: | Ilina, Y, Lorent, C, Katz, S, Jeoung, J.H, Shima, S, Horch, M, Zebger, I, Dobbek, H. | | Deposit date: | 2019-01-12 | | Release date: | 2019-10-23 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.988 Å) | | Cite: | X-ray Crystallography and Vibrational Spectroscopy Reveal the Key Determinants of Biocatalytic Dihydrogen Cycling by [NiFe] Hydrogenases.
Angew.Chem.Int.Ed.Engl., 58, 2019
|
|
6QGR
 
 | | The F420-reducing [NiFe] hydrogenase complex from Methanosarcina barkeri at the Nia-S state | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, (R,R)-2,3-BUTANEDIOL, Coenzyme F420 hydrogenase subunit alpha, ... | | Authors: | Ilina, Y, Lorent, C, Katz, S, Jeoung, J.H, Shima, S, Horch, M, Zebger, I, Dobbek, H. | | Deposit date: | 2019-01-12 | | Release date: | 2019-10-23 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.839 Å) | | Cite: | X-ray Crystallography and Vibrational Spectroscopy Reveal the Key Determinants of Biocatalytic Dihydrogen Cycling by [NiFe] Hydrogenases.
Angew.Chem.Int.Ed.Engl., 58, 2019
|
|
6GAL
 
 | | Structure of fully reduced Hydrogenase (Hyd-1) variant E28Q collected at pH 10 | | Descriptor: | 3-CYCLOHEXYL-1-PROPYLSULFONIC ACID, CHLORIDE ION, FE3-S4 CLUSTER, ... | | Authors: | Carr, S.B, Armstrong, F.A, Evans, R.M. | | Deposit date: | 2018-04-11 | | Release date: | 2019-02-27 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | Mechanistic Exploitation of a Self-Repairing, Blocked Proton Transfer Pathway in an O2-Tolerant [NiFe]-Hydrogenase.
J. Am. Chem. Soc., 140, 2018
|
|
6GAN
 
 | | Structure of fully reduced Hydrogenase (Hyd-2) variant E14Q | | Descriptor: | CARBONMONOXIDE-(DICYANO) IRON, FE3-S4 CLUSTER, Hydrogenase-2 large chain, ... | | Authors: | Carr, S.B, Armstrong, F.A, Evans, R.M. | | Deposit date: | 2018-04-11 | | Release date: | 2019-02-20 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Mechanistic Exploitation of a Self-Repairing, Blocked Proton Transfer Pathway in an O2-Tolerant [NiFe]-Hydrogenase.
J. Am. Chem. Soc., 140, 2018
|
|
6GAM
 
 | | Structure of E14Q variant of E. coli hydrogenase-2 (as-isolated enzyme) | | Descriptor: | CARBONMONOXIDE-(DICYANO) IRON, FE3-S4 CLUSTER, Hydrogenase-2 large chain, ... | | Authors: | Carr, S.B, Armstrong, F.A, Evans, R.M. | | Deposit date: | 2018-04-11 | | Release date: | 2019-02-20 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Mechanistic Exploitation of a Self-Repairing, Blocked Proton Transfer Pathway in an O2-Tolerant [NiFe]-Hydrogenase.
J. Am. Chem. Soc., 140, 2018
|
|
6G94
 
 | |
6G7M
 
 | | Four-site variant (Y222C, C197S, C432S, C433S) of E. coli hydrogenase-2 | | Descriptor: | CARBONMONOXIDE-(DICYANO) IRON, FE3-S4 CLUSTER, Hydrogenase-2 large chain, ... | | Authors: | Carr, S.B, Armstrong, F.A, Zhang, L, Beaton, S.E. | | Deposit date: | 2018-04-06 | | Release date: | 2019-04-03 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.71 Å) | | Cite: | Direct visible light activation of a surface cysteine-engineered [NiFe]-hydrogenase by silver nanoclusters
Energy Environ Sci, 2019
|
|
6G7R
 
 | | Structure of fully reduced variant E28Q of E. coli hydrogenase-1 at pH 8 | | Descriptor: | CHLORIDE ION, DODECYL-BETA-D-MALTOSIDE, FE3-S4 CLUSTER, ... | | Authors: | Carr, S.B, Armstrong, F.A, Evans, R.M. | | Deposit date: | 2018-04-06 | | Release date: | 2019-02-27 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Mechanistic Exploitation of a Self-Repairing, Blocked Proton Transfer Pathway in an O2-Tolerant [NiFe]-Hydrogenase.
J. Am. Chem. Soc., 140, 2018
|
|
6CFW
 
 | |
6FPW
 
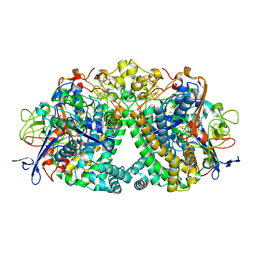 | | Structure of fully reduced Hydrogenase (Hyd-1) | | Descriptor: | CHLORIDE ION, DODECYL-BETA-D-MALTOSIDE, FE3-S4 CLUSTER, ... | | Authors: | Carr, S.B, Armstrong, F.A, Evans, R.M. | | Deposit date: | 2018-02-12 | | Release date: | 2019-02-27 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Mechanistic Exploitation of a Self-Repairing, Blocked Proton Transfer Pathway in an O2-Tolerant [NiFe]-Hydrogenase.
J. Am. Chem. Soc., 140, 2018
|
|
6FPO
 
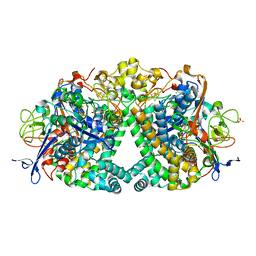 | | High resolution structure of native Hydrogenase (Hyd-1) | | Descriptor: | CARBONMONOXIDE-(DICYANO) IRON, CHLORIDE ION, DODECYL-BETA-D-MALTOSIDE, ... | | Authors: | Carr, S.B, Armstrong, F.A, Evans, R.M. | | Deposit date: | 2018-02-11 | | Release date: | 2019-02-27 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.05 Å) | | Cite: | Mechanistic Exploitation of a Self-Repairing, Blocked Proton Transfer Pathway in an O2-Tolerant [NiFe]-Hydrogenase.
J. Am. Chem. Soc., 140, 2018
|
|
6FPI
 
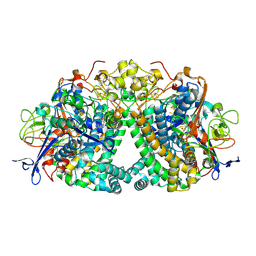 | | Structure of fully reduced Hydrogenase (Hyd-1) variant E28Q | | Descriptor: | CHLORIDE ION, DODECYL-BETA-D-MALTOSIDE, FE3-S4 CLUSTER, ... | | Authors: | Carr, S.B, Armstrong, F.A, Evans, R.M. | | Deposit date: | 2018-02-09 | | Release date: | 2018-12-19 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Mechanistic Exploitation of a Self-Repairing, Blocked Proton Transfer Pathway in an O2-Tolerant [NiFe]-Hydrogenase.
J. Am. Chem. Soc., 140, 2018
|
|
5YY0
 
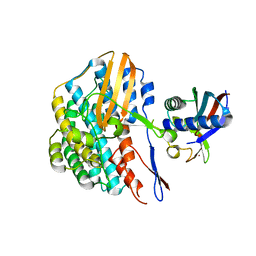 | | Crystal structure of the HyhL-HypA complex (form II) | | Descriptor: | Cytosolic NiFe-hydrogenase, alpha subunit, Probable hydrogenase nickel incorporation protein HypA, ... | | Authors: | Kwon, S, Watanabe, S, Nishitani, Y, Miki, K. | | Deposit date: | 2017-12-07 | | Release date: | 2018-06-20 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (3.243 Å) | | Cite: | Crystal structures of a [NiFe] hydrogenase large subunit HyhL in an immature state in complex with a Ni chaperone HypA.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
5YXY
 
 | | Crystal structure of the HyhL-HypA complex (form I) | | Descriptor: | Cytosolic NiFe-hydrogenase, alpha subunit, Probable hydrogenase nickel incorporation protein HypA, ... | | Authors: | Kwon, S, Watanabe, S, Nishitani, Y, Miki, K. | | Deposit date: | 2017-12-07 | | Release date: | 2018-06-20 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (3.299 Å) | | Cite: | Crystal structures of a [NiFe] hydrogenase large subunit HyhL in an immature state in complex with a Ni chaperone HypA.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
6EN9
 
 | | E. coli Hydrogenase-2 (hydrogen reduced form) | | Descriptor: | CARBONMONOXIDE-(DICYANO) IRON, FE3-S4 CLUSTER, Hydrogenase-2 large chain, ... | | Authors: | Carr, S.B, Beaton, S.E, Evans, R.M, Armstrong, F.A. | | Deposit date: | 2017-10-04 | | Release date: | 2018-04-18 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | The structure of hydrogenase-2 fromEscherichia coli: implications for H2-driven proton pumping.
Biochem. J., 475, 2018
|
|
6EHS
 
 | | E. coli Hydrogenase-2 chemically reduced structure | | Descriptor: | CARBONMONOXIDE-(DICYANO) IRON, DITHIONITE, FE3-S4 CLUSTER, ... | | Authors: | Carr, S.B, Evans, R.M, Beaton, S.E, Armstrong, F.A. | | Deposit date: | 2017-09-15 | | Release date: | 2018-04-18 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | The structure of hydrogenase-2 fromEscherichia coli: implications for H2-driven proton pumping.
Biochem. J., 475, 2018
|
|
6EHQ
 
 | | E. coli Hydrogenase-2 (as isolated form). | | Descriptor: | CARBONMONOXIDE-(DICYANO) IRON, FE3-S4 CLUSTER, Hydrogenase-2 large chain, ... | | Authors: | Carr, S.B, Beaton, S.E, Evans, R.M, Armstrong, F.A. | | Deposit date: | 2017-09-14 | | Release date: | 2018-04-18 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | The structure of hydrogenase-2 fromEscherichia coli: implications for H2-driven proton pumping.
Biochem. J., 475, 2018
|
|
5Y4N
 
 | | Crystal structure of aerobically purified and anaerobically crystallized D. vulgaris Miyazaki F [NiFe]-hydrogenase | | Descriptor: | (4R)-2-METHYLPENTANE-2,4-DIOL, (4S)-2-METHYL-2,4-PENTANEDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, ... | | Authors: | Nishikawa, K, Mochida, S, Hiromoto, T, Shibata, N, Higuchi, Y. | | Deposit date: | 2017-08-04 | | Release date: | 2018-08-08 | | Last modified: | 2020-02-19 | | Method: | X-RAY DIFFRACTION (1.69 Å) | | Cite: | Ni-elimination from the active site of the standard [NiFe]‐hydrogenase upon oxidation by O2.
J.Inorg.Biochem., 177, 2017
|
|
