2PC0
 
 | | Apo Wild-type HIV Protease in the open conformation | | 分子名称: | MAGNESIUM ION, Protease, R-1,2-PROPANEDIOL | | 著者 | Heaslet, H, Rosenfeld, R, Giffin, M.J, Elder, J.H, McRee, D.E, Stout, C.D. | | 登録日 | 2007-03-29 | | 公開日 | 2007-06-26 | | 最終更新日 | 2023-08-30 | | 実験手法 | X-RAY DIFFRACTION (1.4 Å) | | 主引用文献 | Conformational flexibility in the flap domains of ligand-free HIV protease.
Acta Crystallogr.,Sect.D, 63, 2007
|
|
3DCR
 
 | | X-ray structure of HIV-1 protease and hydrated form of ketomethylene isostere inhibitor | | 分子名称: | Chemical analogue HIV-1 protease, N~2~-[(2R,5S)-5-({(2S,3S)-2-[(N-acetyl-L-threonyl)amino]-3-methylpent-4-enoyl}amino)-2-butyl-4,4-dihydroxynonanoyl]-L-glutaminyl-L-argininamide | | 著者 | Torbeev, V.Y, Mandal, K, Terechko, V.A, Kent, S.B.H. | | 登録日 | 2008-06-04 | | 公開日 | 2008-08-19 | | 最終更新日 | 2023-11-15 | | 実験手法 | X-RAY DIFFRACTION (1.4 Å) | | 主引用文献 | Crystal structure of chemically synthesized HIV-1 protease and a ketomethylene isostere inhibitor based on the p2/NC cleavage site
Bioorg.Med.Chem.Lett., 18, 2008
|
|
3KA2
 
 | |
4DJP
 
 | | Crystal Structure of wild-type HIV-1 Protease in Complex with MKP73 | | 分子名称: | PHOSPHATE ION, Pol polyprotein, methyl (2S)-3-({[(2S,3R)-3-hydroxy-4-{[(4-methoxyphenyl)sulfonyl][(2S)-2-methylbutyl]amino}-1-phenylbutan-2-yl]carbamoyl}oxy)-2-methylpropanoate | | 著者 | Schiffer, C.A, Nalam, M.N.L. | | 登録日 | 2012-02-02 | | 公開日 | 2012-08-01 | | 最終更新日 | 2024-02-28 | | 実験手法 | X-RAY DIFFRACTION (1.4 Å) | | 主引用文献 | Design, synthesis, and biological and structural evaluations of novel HIV-1 protease inhibitors to combat drug resistance.
J.Med.Chem., 55, 2012
|
|
4DJQ
 
 | | Crystal Structure of wild-type HIV-1 Protease in Complex with MKP86 | | 分子名称: | 2-(2-oxoimidazolidin-1-yl)ethyl [(2S,3R)-3-hydroxy-4-{[(4-methoxyphenyl)sulfonyl][(2S)-2-methylbutyl]amino}-1-phenylbutan-2-yl]carbamate, PHOSPHATE ION, Pol polyprotein | | 著者 | Schiffer, C.A, Nalam, M.N.L. | | 登録日 | 2012-02-02 | | 公開日 | 2012-08-01 | | 最終更新日 | 2024-02-28 | | 実験手法 | X-RAY DIFFRACTION (1.4 Å) | | 主引用文献 | Design, synthesis, and biological and structural evaluations of novel HIV-1 protease inhibitors to combat drug resistance.
J.Med.Chem., 55, 2012
|
|
3QAA
 
 | | HIV-1 wild type protease with a substituted bis-Tetrahydrofuran inhibitor, GRL-044-10A | | 分子名称: | (3R,3aS,4R,6aR)-4-methoxyhexahydrofuro[2,3-b]furan-3-yl [(2S,3R)-3-hydroxy-4-{[(4-methoxyphenyl)sulfonyl](2-methylpropyl)amino}-1-phenylbutan-2-yl]carbamate, CHLORIDE ION, GLYCEROL, ... | | 著者 | Wang, Y.-F, Agniswamy, J, Weber, I.T. | | 登録日 | 2011-01-10 | | 公開日 | 2011-12-21 | | 最終更新日 | 2023-09-13 | | 実験手法 | X-RAY DIFFRACTION (1.4 Å) | | 主引用文献 | Design of substituted bis-Tetrahydrofuran (bis-THF)-derived Potent HIV-1 Protease Inhibitors, Protein-ligand X-ray Structure, and Convenient Syntheses of bis-THF and Substituted bis-THF Ligands.
ACS Med Chem Lett, 2, 2011
|
|
1SDV
 
 | | Crystal structures of HIV protease V82A and L90M mutants reveal changes in indinavir binding site. | | 分子名称: | CHLORIDE ION, N-[2(R)-HYDROXY-1(S)-INDANYL]-5-[(2(S)-TERTIARY BUTYLAMINOCARBONYL)-4(3-PYRIDYLMETHYL)PIPERAZINO]-4(S)-HYDROXY-2(R)-PHENYLMETHYLPENTANAMIDE, protease RETROPEPSIN | | 著者 | Mahalingam, B, Wang, Y.-F, Boross, P.I, Tozser, J, Louis, J.M, Harrison, R.W, Weber, I.T. | | 登録日 | 2004-02-14 | | 公開日 | 2004-05-25 | | 最終更新日 | 2023-08-23 | | 実験手法 | X-RAY DIFFRACTION (1.4 Å) | | 主引用文献 | Crystal structures of HIV protease V82A and L90M
mutants reveal changes in the indinavir-binding site
Eur.J.Biochem., 271, 2004
|
|
4FM6
 
 | |
3SLZ
 
 | | The crystal structure of XMRV protease complexed with TL-3 | | 分子名称: | FORMIC ACID, SODIUM ION, benzyl [(1S,4S,7S,8R,9R,10S,13S,16S)-7,10-dibenzyl-8,9-dihydroxy-1,16-dimethyl-4,13-bis(1-methylethyl)-2,5,12,15,18-pentaoxo-20-phenyl-19-oxa-3,6,11,14,17-pentaazaicos-1-yl]carbamate, ... | | 著者 | Li, M, Gustchina, A, Wlodawer, A. | | 登録日 | 2011-06-27 | | 公開日 | 2011-10-12 | | 最終更新日 | 2024-02-28 | | 実験手法 | X-RAY DIFFRACTION (1.4 Å) | | 主引用文献 | Structural and biochemical characterization of the inhibitor complexes of xenotropic murine leukemia virus-related virus protease.
Febs J., 278, 2011
|
|
2AOD
 
 | | Crystal structure analysis of HIV-1 protease with a substrate analog P2-NC | | 分子名称: | DIMETHYL SULFOXIDE, GLYCEROL, HIV-1 PROTEASE, ... | | 著者 | Tie, Y, Boross, P.I, Wang, Y.F, Gaddis, L, Liu, F, Chen, X, Tozser, J, Harrison, R.W, Weber, I.T. | | 登録日 | 2005-08-12 | | 公開日 | 2006-01-17 | | 最終更新日 | 2024-03-13 | | 実験手法 | X-RAY DIFFRACTION (1.4 Å) | | 主引用文献 | Molecular basis for substrate recognition and drug resistance from 1.1 to 1.6 angstroms resolution crystal structures of HIV-1 protease mutants with substrate analogs.
Febs J., 272, 2005
|
|
2AOI
 
 | | Crystal structure analysis of HIV-1 protease with a substrate analog P1-P6 | | 分子名称: | PEPTIDE INHIBITOR, POL POLYPROTEIN, SULFATE ION | | 著者 | Tie, Y, Boross, P.I, Wang, Y.F, Gaddis, L, Liu, F, Chen, X, Tozser, J, Harrison, R.W, Weber, I.T. | | 登録日 | 2005-08-12 | | 公開日 | 2006-01-17 | | 最終更新日 | 2023-08-23 | | 実験手法 | X-RAY DIFFRACTION (1.4 Å) | | 主引用文献 | Molecular basis for substrate recognition and drug resistance from 1.1 to 1.6 angstroms resolution crystal structures of HIV-1 protease mutants with substrate analogs.
Febs J., 272, 2005
|
|
2QNP
 
 | | HIV-1 Protease in complex with a iodo decorated pyrrolidine-based inhibitor | | 分子名称: | CHLORIDE ION, Gag-Pol polyprotein (Pr160Gag-Pol), N,N'-(3S,4S)-pyrrolidine-3,4-diylbis[N-(4-iodobenzyl)benzenesulfonamide] | | 著者 | Boettcher, J, Blum, A, Heine, A, Diederich, W.E, Klebe, G. | | 登録日 | 2007-07-19 | | 公開日 | 2008-04-15 | | 最終更新日 | 2024-03-13 | | 実験手法 | X-RAY DIFFRACTION (1.41 Å) | | 主引用文献 | Structure-Guided Design of C2-Symmetric HIV-1 Protease Inhibitors Based on a Pyrrolidine Scaffold.
J.Med.Chem., 51, 2008
|
|
6OGQ
 
 | | X-ray crystal structure of darunavir-resistant HIV-1 protease (P30) in complex with GRL-003 | | 分子名称: | (3S,3aR,5R,7aS,8S)-hexahydro-4H-3,5-methanofuro[2,3-b]pyran-8-yl [(2S,3R)-4-[{[2-(cyclopropylamino)-1,3-benzothiazol-6-yl]sulfonyl}(2-methylpropyl)amino]-1-(4-fluorophenyl)-3-hydroxybutan-2-yl]carbamate, DI(HYDROXYETHYL)ETHER, GLYCEROL, ... | | 著者 | Bulut, H, Hattori, S.I, Aoki-Ogata, H, Hayashi, H, Aoki, M, Ghosh, A.K, Mitsuya, H. | | 登録日 | 2019-04-03 | | 公開日 | 2020-04-08 | | 最終更新日 | 2023-10-11 | | 実験手法 | X-RAY DIFFRACTION (1.41 Å) | | 主引用文献 | Single atom changes in newly synthesized HIV protease inhibitors reveal structural basis for extreme affinity, high genetic barrier, and adaptation to the HIV protease plasticity.
Sci Rep, 10, 2020
|
|
2AOH
 
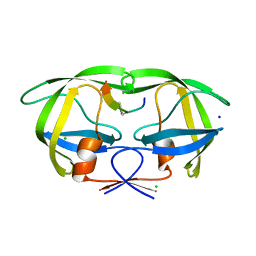 | | Crystal structure analysis of HIV-1 Protease mutant V82A with a substrate analog P6-PR | | 分子名称: | CHLORIDE ION, PEPTIDE INHIBITOR, POL POLYPROTEIN, ... | | 著者 | Tie, Y, Boross, P.I, Wang, Y.F, Gaddis, L, Liu, F, Chen, X, Tozser, J, Harrison, R.W, Weber, I.T. | | 登録日 | 2005-08-12 | | 公開日 | 2006-01-17 | | 最終更新日 | 2023-08-23 | | 実験手法 | X-RAY DIFFRACTION (1.42 Å) | | 主引用文献 | Molecular basis for substrate recognition and drug resistance from 1.1 to 1.6 angstroms resolution crystal structures of HIV-1 protease mutants with substrate analogs.
Febs J., 272, 2005
|
|
3S54
 
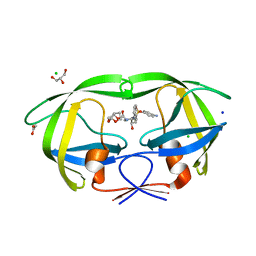 | | HIV-1 protease triple mutants V32I, I47V, V82I with antiviral drug darunavir in space group P21212 | | 分子名称: | (3R,3AS,6AR)-HEXAHYDROFURO[2,3-B]FURAN-3-YL(1S,2R)-3-[[(4-AMINOPHENYL)SULFONYL](ISOBUTYL)AMINO]-1-BENZYL-2-HYDROXYPROPYLCARBAMATE, ACETATE ION, CHLORIDE ION, ... | | 著者 | Tie, Y.-F, Wang, Y.-F, Weber, I.T. | | 登録日 | 2011-05-20 | | 公開日 | 2012-03-21 | | 最終更新日 | 2023-09-13 | | 実験手法 | X-RAY DIFFRACTION (1.42 Å) | | 主引用文献 | Critical differences in HIV-1 and HIV-2 protease specificity for clinical inhibitors.
Protein Sci., 21, 2012
|
|
3VFA
 
 | | Crystal Structure of HIV-1 Protease Mutant V82A with novel P1'-Ligands GRL-02031 | | 分子名称: | (3aS,5R,6aR)-hexahydro-2H-cyclopenta[b]furan-5-yl [(1S,2R)-1-benzyl-2-hydroxy-3-([(4-methoxyphenyl)sulfonyl]{[(2R)-5-oxopyrrolidin-2-yl]methyl}amino)propyl]carbamate, CHLORIDE ION, SODIUM ION, ... | | 著者 | Yu, X.X, Wang, Y.F, Chang, Y.C.E, Weber, I.T. | | 登録日 | 2012-01-09 | | 公開日 | 2012-11-21 | | 最終更新日 | 2023-09-13 | | 実験手法 | X-RAY DIFFRACTION (1.43 Å) | | 主引用文献 | Potent antiviral HIV-1 protease inhibitor GRL-02031 adapts to the structures of drug resistant mutants with its P1'-pyrrolidinone ring.
J.Med.Chem., 55, 2012
|
|
5AH9
 
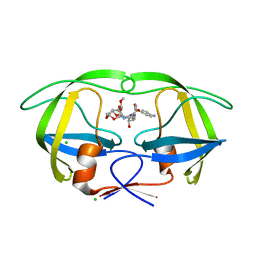 | | Disubstituted bis-THF moieties as new P2 ligands in non-peptidal HIV- 1 Protease Inhibitors (II) | | 分子名称: | (3R,3aS,4R,6aR)-4-(2-methoxyethoxy)hexahydrofuro[2,3-b]furan-3-yl [(2S,3R)-4-{[(4-aminophenyl)sulfonyl](2-methylpropyl)amino}-3-hydroxy-1-phenylbutan-2-yl]carbamate, CHLORIDE ION, PROTEASE | | 著者 | Hohlfeld, K, Wegner, J.K, Kesteleyn, B, Linclau, B, Enstrom, O, Unge, J. | | 登録日 | 2015-02-05 | | 公開日 | 2015-05-06 | | 最終更新日 | 2024-05-01 | | 実験手法 | X-RAY DIFFRACTION (1.44 Å) | | 主引用文献 | Disubstituted Bis-Thf Moieties as New P2 Ligands in Non-Peptidal HIV-1 Protease Inhibitors (II).
J.Med.Chem., 58, 2015
|
|
2FNT
 
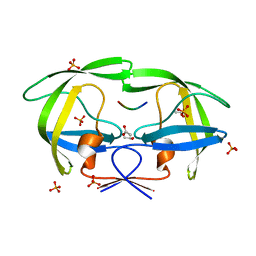 | | Crystal structure of a drug-resistant (V82A) inactive (D25N) HIV-1 protease complexed with AP2V variant of HIV-1 NC-p1 substrate. | | 分子名称: | ACETATE ION, NC-p1 substrate PEPTIDE, PHOSPHATE ION, ... | | 著者 | Prabu-Jeyabaln, M, Nalivaika, E.A, Schiffer, C.A. | | 登録日 | 2006-01-11 | | 公開日 | 2006-09-05 | | 最終更新日 | 2023-08-30 | | 実験手法 | X-RAY DIFFRACTION (1.44 Å) | | 主引用文献 | Mechanism of substrate recognition by drug-resistant human immunodeficiency virus type 1 protease variants revealed by a novel structural intermediate.
J.Virol., 80, 2006
|
|
3NYG
 
 | |
3O9I
 
 | | Crystal Structure of wild-type HIV-1 Protease in complex with af61 | | 分子名称: | (3R,3aS,6aR)-hexahydrofuro[2,3-b]furan-3-yl {(1S,2R)-3-[(1,3-benzothiazol-6-ylsulfonyl)(2-ethylbutyl)amino]-1-benzyl-2-hydroxypropyl}carbamate, PHOSPHATE ION, Protease | | 著者 | Schiffer, C.A, Nalam, M.N.L. | | 登録日 | 2010-08-04 | | 公開日 | 2011-08-10 | | 最終更新日 | 2024-04-03 | | 実験手法 | X-RAY DIFFRACTION (1.45 Å) | | 主引用文献 | Substrate envelope-designed potent HIV-1 protease inhibitors to avoid drug resistance.
Chem.Biol., 20, 2013
|
|
3UFN
 
 | | Crystal Structure of Multidrug Resistant HIV-1 Protease Clinical Isolate PR20 in Complex with Saquinavir | | 分子名称: | (2S)-N-[(2S,3R)-4-[(2S,3S,4aS,8aS)-3-(tert-butylcarbamoyl)-3,4,4a,5,6,7,8,8a-octahydro-1H-isoquinolin-2-yl]-3-hydroxy-1 -phenyl-butan-2-yl]-2-(quinolin-2-ylcarbonylamino)butanediamide, CHLORIDE ION, HIV-1 protease | | 著者 | Agniswamy, J, Chen-Hsiang, S, Aniana, A, Sayer, J.M, Louis, J.M, Weber, I.T. | | 登録日 | 2011-11-01 | | 公開日 | 2012-03-28 | | 最終更新日 | 2024-02-28 | | 実験手法 | X-RAY DIFFRACTION (1.45 Å) | | 主引用文献 | HIV-1 protease with 20 mutations exhibits extreme resistance to clinical inhibitors through coordinated structural rearrangements.
Biochemistry, 51, 2012
|
|
4Z50
 
 | | Crystal Structure of Multidrug Resistant HIV-1 Protease Clinical Isolate PR20D25N with Tucked Flap | | 分子名称: | CHLORIDE ION, GLYCEROL, Protease, ... | | 著者 | Agniswamy, J, Shen, C.-H, Weber, I.T. | | 登録日 | 2015-04-02 | | 公開日 | 2015-10-14 | | 最終更新日 | 2023-09-27 | | 実験手法 | X-RAY DIFFRACTION (1.45 Å) | | 主引用文献 | Conformational variation of an extreme drug resistant mutant of HIV protease.
J.Mol.Graph.Model., 62, 2015
|
|
2PK6
 
 | | Crystal Structure of HIV-1 Protease (Q7K, L33I, L63I) in Complex with KNI-10033 | | 分子名称: | (4R)-N-[(1S,2R)-2-hydroxy-2,3-dihydro-1H-inden-1-yl]-3-[(2S,3S)-2-hydroxy-3-({N-[(isoquinolin-5-yloxy)acetyl]-S-methyl- L-cysteinyl}amino)-4-phenylbutanoyl]-5,5-dimethyl-1,3-thiazolidine-4-carboxamide, GLYCEROL, Protease | | 著者 | Armstrong, A.A, Lafont, V, Kiso, Y, Freire, E, Amzel, L.M. | | 登録日 | 2007-04-17 | | 公開日 | 2007-05-08 | | 最終更新日 | 2023-08-30 | | 実験手法 | X-RAY DIFFRACTION (1.45 Å) | | 主引用文献 | Compensating enthalpic and entropic changes hinder binding affinity optimization.
Chem.Biol.Drug Des., 69, 2007
|
|
3PWR
 
 | | HIV-1 Protease Mutant L76V complexed with Saquinavir | | 分子名称: | (2S)-N-[(2S,3R)-4-[(2S,3S,4aS,8aS)-3-(tert-butylcarbamoyl)-3,4,4a,5,6,7,8,8a-octahydro-1H-isoquinolin-2-yl]-3-hydroxy-1 -phenyl-butan-2-yl]-2-(quinolin-2-ylcarbonylamino)butanediamide, CHLORIDE ION, GLYCEROL, ... | | 著者 | Zhang, Y, Weber, I.T. | | 登録日 | 2010-12-08 | | 公開日 | 2011-04-20 | | 最終更新日 | 2023-09-13 | | 実験手法 | X-RAY DIFFRACTION (1.45 Å) | | 主引用文献 | The L76V Drug Resistance Mutation Decreases the Dimer Stability and Rate of Autoprocessing of HIV-1 Protease by Reducing Internal Hydrophobic Contacts.
Biochemistry, 50, 2011
|
|
3QBF
 
 | |
