8QKT
 
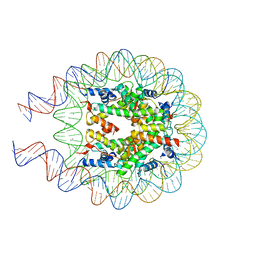 | |
8WDV
 
 | | Photosynthetic LH1-RC complex from the purple sulfur bacterium Allochromatium vinosum purified by Ca2+-DEAE | | Descriptor: | (1R)-2-{[{[(2S)-2,3-DIHYDROXYPROPYL]OXY}(HYDROXY)PHOSPHORYL]OXY}-1-[(PALMITOYLOXY)METHYL]ETHYL (11E)-OCTADEC-11-ENOATE, (2S)-3-hydroxypropane-1,2-diyl dihexadecanoate, Antenna complex alpha/beta subunit, ... | | Authors: | Tani, K, Kanno, R, Harada, A, Kobayashi, A, Minamino, A, Nakamura, N, Ji, X.-C, Purba, E.R, Hall, M, Yu, L.-J, Madigan, M.T, Mizoguchi, A, Iwasaki, K, Humbel, B.M, Kimura, Y, Wang-Otomo, Z.-Y. | | Deposit date: | 2023-09-16 | | Release date: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (2.24 Å) | | Cite: | High-resolution structure and biochemical properties of the LH1-RC photocomplex from the model purple sulfur bacterium, Allochromatium vinosum
Commun Biol, 7, 2024
|
|
8WDU
 
 | | Photosynthetic LH1-RC complex from the purple sulfur bacterium Allochromatium vinosum purified by sucrose density | | Descriptor: | (1R)-2-{[{[(2S)-2,3-DIHYDROXYPROPYL]OXY}(HYDROXY)PHOSPHORYL]OXY}-1-[(PALMITOYLOXY)METHYL]ETHYL (11E)-OCTADEC-11-ENOATE, (2S)-3-hydroxypropane-1,2-diyl dihexadecanoate, Antenna complex alpha/beta subunit, ... | | Authors: | Tani, K, Kanno, R, Harada, A, Kobayashi, A, Minamino, A, Nakamura, N, Ji, X.-C, Purba, E.R, Hall, M, Yu, L.-J, Madigan, M.T, Mizoguchi, A, Iwasaki, K, Humbel, B.M, Kimura, Y, Wang-Otomo, Z.-Y. | | Deposit date: | 2023-09-16 | | Release date: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (2.24 Å) | | Cite: | High-resolution structure and biochemical properties of the LH1-RC photocomplex from the model purple sulfur bacterium, Allochromatium vinosum
Commun Biol, 7, 2024
|
|
8WDI
 
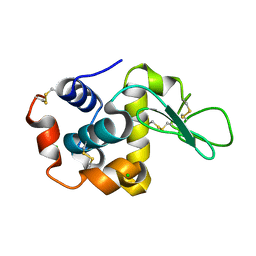 | |
8U7M
 
 | | Human retinal variant phosphomimetic IMPDH1(595)-S477D free octamer bound by GTP, ATP, IMP, and NAD+ | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, GUANOSINE-5'-TRIPHOSPHATE, INOSINIC ACID, ... | | Authors: | Calise, S.J, Kollman, J.M. | | Deposit date: | 2023-09-15 | | Release date: | 2024-01-31 | | Last modified: | 2024-02-14 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Light-sensitive phosphorylation regulates retinal IMPDH1 activity and filament assembly.
J.Cell Biol., 223, 2024
|
|
8U7V
 
 | | Human retinal variant phosphomimetic IMPDH1(546)-S477D filament bound by GTP, ATP, IMP, and NAD+, interface-centered | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, GUANOSINE-5'-TRIPHOSPHATE, INOSINIC ACID, ... | | Authors: | Calise, S.J, Kollman, J.M. | | Deposit date: | 2023-09-15 | | Release date: | 2024-01-31 | | Last modified: | 2024-02-14 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Light-sensitive phosphorylation regulates retinal IMPDH1 activity and filament assembly.
J.Cell Biol., 223, 2024
|
|
8U7Q
 
 | | Human retinal variant phosphomimetic IMPDH1(546)-S477D filament bound by GTP, ATP, IMP, and NAD+, octamer-centered | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, GUANOSINE-5'-TRIPHOSPHATE, INOSINIC ACID, ... | | Authors: | Calise, S.J, Kollman, J.M. | | Deposit date: | 2023-09-15 | | Release date: | 2024-01-31 | | Last modified: | 2024-02-14 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Light-sensitive phosphorylation regulates retinal IMPDH1 activity and filament assembly.
J.Cell Biol., 223, 2024
|
|
8U5Z
 
 | | Structure of Mango II variant aptamer bound to T01-7M-B | | Descriptor: | 2-[(E)-(1,7-dimethylquinolin-4(1H)-ylidene)methyl]-3-{2,16-dioxo-20-[(3aR,4S,6aR)-2-oxohexahydro-1H-thieno[3,4-d]imidazol-4-yl]-6,9,12-trioxa-3,15-diazaicosan-1-yl}-1,3-benzothiazol-3-ium, Mango II variant, POTASSIUM ION, ... | | Authors: | Passalacqua, L.F.M, Ferre-D'Amare, A.R. | | Deposit date: | 2023-09-13 | | Release date: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structure of Mango II variant aptamer bound to T01-7M-B
To Be Published
|
|
8WCQ
 
 | | Cryo-EM structure of nanodisc (PE:PS:PC) reconstituted GLIC at pH 4 in intermediate state | | Descriptor: | 1,2-dioleoyl-sn-glycero-3-phosphoethanolamine, CHLORIDE ION, Proton-gated ion channel | | Authors: | Bharambe, N, Li, Z, Basak, S. | | Deposit date: | 2023-09-13 | | Release date: | 2024-04-17 | | Method: | ELECTRON MICROSCOPY (3.35 Å) | | Cite: | Cryo-EM structures of prokaryotic ligand-gated ion channel GLIC provide insights into gating in a lipid environment.
Nat Commun, 15, 2024
|
|
8WCR
 
 | | Cryo-EM structure of nanodisc (PE:PS:PC) reconstituted GLIC at pH 4 in open state | | Descriptor: | 1,2-dioleoyl-sn-glycero-3-phosphoethanolamine, CHLORIDE ION, Proton-gated ion channel | | Authors: | Bharambe, N, Li, Z, Basak, S. | | Deposit date: | 2023-09-13 | | Release date: | 2024-04-17 | | Method: | ELECTRON MICROSCOPY (2.74 Å) | | Cite: | Cryo-EM structures of prokaryotic ligand-gated ion channel GLIC provide insights into gating in a lipid environment.
Nat Commun, 15, 2024
|
|
8U5F
 
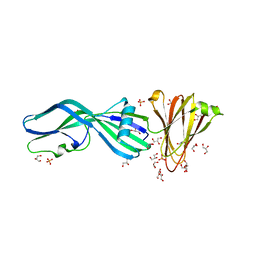 | | Crystal Structure of Trypsinized Clostridium perfringens Enterotoxin | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, GLYCEROL, Heat-labile enterotoxin B chain, ... | | Authors: | Kapoor, S, Ogbu, C.P, Vecchio, A.J. | | Deposit date: | 2023-09-12 | | Release date: | 2023-09-27 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.32 Å) | | Cite: | Structural Basis of Clostridium perfringens Enterotoxin Activation and Oligomerization by Trypsin.
Toxins, 15, 2023
|
|
8U5T
 
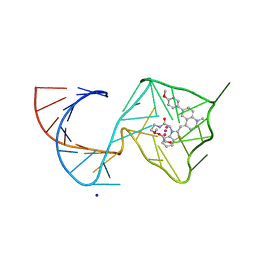 | | Structure of Mango II variant aptamer bound to T01-6A-B | | Descriptor: | 3-{2,16-dioxo-20-[(3aS,4R,6aS)-2-oxohexahydro-1H-thieno[3,4-d]imidazol-4-yl]-6,9,12-trioxa-3,15-diazaicosan-1-yl}-2-{(E)-[6-(4-methoxyphenyl)-1-methylquinolin-4(1H)-ylidene]methyl}-1,3-benzothiazol-3-ium, Mango II variant, POTASSIUM ION, ... | | Authors: | Passalacqua, L.F.M, Ferre-D'Amare, A.R. | | Deposit date: | 2023-09-12 | | Release date: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure of Mango II variant aptamer bound to T01-6A-B
To Be Published
|
|
8U5R
 
 | |
8U4A
 
 | |
8U4F
 
 | |
8U49
 
 | | The Apo Crystal Structure of BlCel9A from Glycoside Hydrolase Family 9 | | Descriptor: | 1,2-ETHANEDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CALCIUM ION, ... | | Authors: | Araujo, E.A, Polikarpov, I. | | Deposit date: | 2023-09-10 | | Release date: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Molecular mechanism of cellulose depolymerization by the two-domain BlCel9A enzyme from the glycoside hydrolase family 9.
Carbohydr Polym, 329, 2024
|
|
8QHK
 
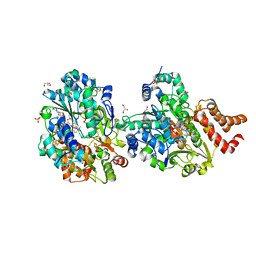 | | Crystal structure of reduced respiratory Complex I subunits NuoEF from Aquifex aeolicus bound to reduced 3-acetylpyridine adenine dinucleotide | | Descriptor: | 1-DEOXY-1-(7,8-DIMETHYL-2,4-DIOXO-3,4-DIHYDRO-2H-BENZO[G]PTERIDIN-1-ID-10(5H)-YL)-5-O-PHOSPHONATO-D-RIBITOL, ACETYL PYRIDINE ADENINE DINUCLEOTIDE, REDUCED, ... | | Authors: | Wohlwend, D, Friedrich, T, Bucka, S. | | Deposit date: | 2023-09-08 | | Release date: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structures of 3-acetylpyridine adenine dinucleotide and ADP-ribose bound to the electron input module of respiratory complex I.
Structure, 2024
|
|
8WAS
 
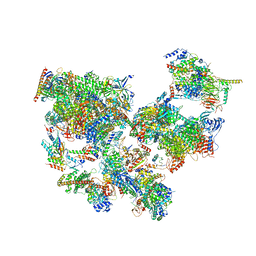 | | Structure of transcribing complex 9 (TC9), the initially transcribing complex with Pol II positioned 9nt downstream of TSS. | | Descriptor: | Alpha-amanitin, CDK-activating kinase assembly factor MAT1, DNA-directed RNA polymerase II subunit E, ... | | Authors: | Chen, X, Liu, W, Wang, Q, Wang, X, Ren, Y, Qu, X, Li, W, Xu, Y. | | Deposit date: | 2023-09-08 | | Release date: | 2023-12-06 | | Last modified: | 2024-01-03 | | Method: | ELECTRON MICROSCOPY (6.13 Å) | | Cite: | Structural visualization of transcription initiation in action.
Science, 382, 2023
|
|
8WAQ
 
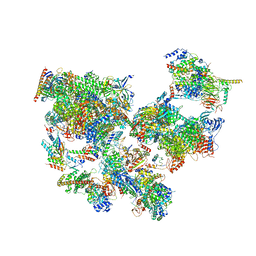 | | Structure of transcribing complex 7 (TC7), the initially transcribing complex with Pol II positioned 7nt downstream of TSS. | | Descriptor: | Alpha-amanitin, CDK-activating kinase assembly factor MAT1, DNA-directed RNA polymerase II subunit E, ... | | Authors: | Chen, X, Liu, W, Wang, Q, Wang, X, Ren, Y, Qu, X, Li, W, Xu, Y. | | Deposit date: | 2023-09-08 | | Release date: | 2023-12-06 | | Last modified: | 2024-01-03 | | Method: | ELECTRON MICROSCOPY (6.29 Å) | | Cite: | Structural visualization of transcription initiation in action.
Science, 382, 2023
|
|
8WAP
 
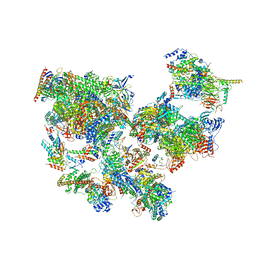 | | Structure of transcribing complex 6 (TC6), the initially transcribing complex with Pol II positioned 6nt downstream of TSS. | | Descriptor: | Alpha-amanitin, CDK-activating kinase assembly factor MAT1, DNA-directed RNA polymerase II subunit E, ... | | Authors: | Chen, X, Liu, W, Wang, Q, Wang, X, Ren, Y, Qu, X, Li, W, Xu, Y. | | Deposit date: | 2023-09-08 | | Release date: | 2023-12-06 | | Last modified: | 2024-01-03 | | Method: | ELECTRON MICROSCOPY (5.85 Å) | | Cite: | Structural visualization of transcription initiation in action.
Science, 382, 2023
|
|
8WAR
 
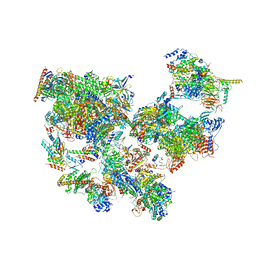 | | Structure of transcribing complex 8 (TC8), the initially transcribing complex with Pol II positioned 8nt downstream of TSS. | | Descriptor: | Alpha-amanitin, CDK-activating kinase assembly factor MAT1, DNA-directed RNA polymerase II subunit E, ... | | Authors: | Chen, X, Liu, W, Wang, Q, Wang, X, Ren, Y, Qu, X, Li, W, Xu, Y. | | Deposit date: | 2023-09-08 | | Release date: | 2023-12-06 | | Last modified: | 2024-01-03 | | Method: | ELECTRON MICROSCOPY (7.2 Å) | | Cite: | Structural visualization of transcription initiation in action.
Science, 382, 2023
|
|
8U36
 
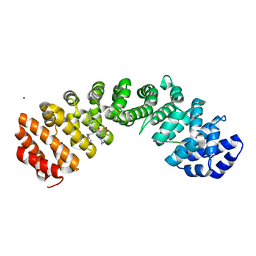 | |
8WA5
 
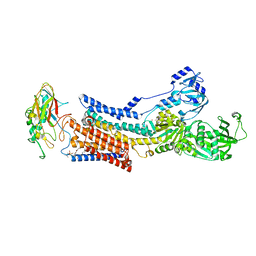 | |
8WAL
 
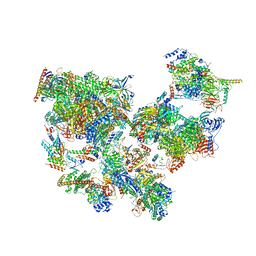 | | Structure of transcribing complex 3 (TC3), the initially transcribing complex with Pol II positioned 3nt downstream of TSS. | | Descriptor: | Alpha-amanitin, CDK-activating kinase assembly factor MAT1, DNA-directed RNA polymerase II subunit E, ... | | Authors: | Chen, X, Liu, W, Wang, Q, Wang, X, Ren, Y, Qu, X, Li, W, Xu, Y. | | Deposit date: | 2023-09-07 | | Release date: | 2023-12-06 | | Last modified: | 2024-01-03 | | Method: | ELECTRON MICROSCOPY (8.52 Å) | | Cite: | Structural visualization of transcription initiation in action.
Science, 382, 2023
|
|
8WAK
 
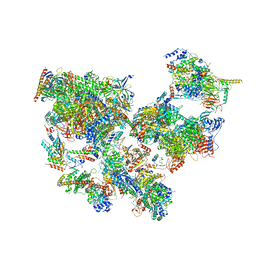 | | Structure of transcribing complex 2 (TC2), the initially transcribing complex with Pol II positioned 2nt downstream of TSS. | | Descriptor: | Alpha-amanitin, CDK-activating kinase assembly factor MAT1, DNA-directed RNA polymerase II subunit E, ... | | Authors: | Chen, X, Liu, W, Wang, Q, Wang, X, Ren, Y, Qu, X, Li, W, Xu, Y. | | Deposit date: | 2023-09-07 | | Release date: | 2023-12-06 | | Last modified: | 2024-01-03 | | Method: | ELECTRON MICROSCOPY (5.47 Å) | | Cite: | Structural visualization of transcription initiation in action.
Science, 382, 2023
|
|
