6CJH
 
 | | Co-crystal structure of MNK2 in complex with an inhibitor | | Descriptor: | 3-phenyl-5-(pyridin-4-yl)-1H-indazole, MAP kinase-interacting serine/threonine-protein kinase 2, ZINC ION | | Authors: | Han, Q. | | Deposit date: | 2018-02-26 | | Release date: | 2018-05-09 | | Method: | X-RAY DIFFRACTION (3.6 Å) | | Cite: | Structure-based Design of Pyridone-Aminal eFT508 Targeting Dysregulated Translation by Selective Mitogen-activated Protein Kinase Interacting Kinases 1 and 2 (MNK1/2) Inhibition.
J. Med. Chem., 61, 2018
|
|
4NV2
 
 | | C50A mutant of Synechococcus VKOR, C2221 crystal form | | Descriptor: | UBIQUINONE-10, VKORC1/thioredoxin domain protein | | Authors: | Liu, S, Cheng, W, Fowle Grider, R, Shen, G, Li, W. | | Deposit date: | 2013-12-04 | | Release date: | 2014-02-12 | | Method: | X-RAY DIFFRACTION (3.61 Å) | | Cite: | Structures of an intramembrane vitamin K epoxide reductase homolog reveal control mechanisms for electron transfer.
Nat Commun, 5, 2014
|
|
4BIU
 
 | | Crystal structure of CpxAHDC (orthorhombic form 1) | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, SENSOR PROTEIN CPXA, SULFATE ION | | Authors: | Mechaly, A.E, Sassoon, N, Betton, J.M, Alzari, P.M. | | Deposit date: | 2013-04-13 | | Release date: | 2014-02-12 | | Method: | X-RAY DIFFRACTION (3.65 Å) | | Cite: | Segmental Helical Motions and Dynamical Asymmetry Modulate Histidine Kinase Autophosphorylation.
Plos Biol., 12, 2014
|
|
6EYC
 
 | | Re-refinement of the MCM2-7 double hexamer using ISOLDE | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, DNA replication licensing factor MCM2, DNA replication licensing factor MCM3, ... | | Authors: | Croll, T.I. | | Deposit date: | 2017-11-11 | | Release date: | 2018-06-20 | | Last modified: | 2019-12-11 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | ISOLDE: a physically realistic environment for model building into low-resolution electron-density maps.
Acta Crystallogr D Struct Biol, 74, 2018
|
|
4EIW
 
 | | Whole cytosolic region of atp-dependent metalloprotease FtsH (G399L) | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ATP-dependent zinc metalloprotease FtsH | | Authors: | Suno, R, Niwa, H, Tsuchiya, D, Yoshida, M, Morikawa, K. | | Deposit date: | 2012-04-06 | | Release date: | 2012-06-06 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (3.9 Å) | | Cite: | Structure of the whole cytosolic region of ATP-dependent protease FtsH
Mol.Cell, 22, 2006
|
|
4FGU
 
 | | Crystal structure of prolegumain | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Legumain | | Authors: | Dall, E, Brandstetter, H. | | Deposit date: | 2012-06-05 | | Release date: | 2013-07-03 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (3.9 Å) | | Cite: | Mechanistic and structural studies on legumain explain its zymogenicity, distinct activation pathways, and regulation.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
5UCG
 
 | | Structure of the PP2C Phosphatase Domain and a Fragment of the Regulatory Domain of the Cell Fate Determinant SpoIIE from Bacillus Subtilis | | Descriptor: | Stage II sporulation protein E | | Authors: | Bradshaw, N, Levdikov, V, Zimanyi, C, Gaudet, R, Wilkinson, A, Losick, R. | | Deposit date: | 2016-12-22 | | Release date: | 2017-05-31 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (3.906 Å) | | Cite: | A widespread family of serine/threonine protein phosphatases shares a common regulatory switch with proteasomal proteases.
Elife, 6, 2017
|
|
7BL1
 
 | | human complex II-BATS bound to membrane-attached Rab5a-GTP | | Descriptor: | Beclin-1, GUANOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Tremel, S, Morado, D.R, Kovtun, O, Williams, R.L, Briggs, J.A.G, Munro, S, Ohashi, Y, Bertram, J, Perisic, O. | | Deposit date: | 2021-01-17 | | Release date: | 2021-03-03 | | Last modified: | 2021-03-24 | | Method: | ELECTRON MICROSCOPY (9.8 Å) | | Cite: | Structural basis for VPS34 kinase activation by Rab1 and Rab5 on membranes.
Nat Commun, 12, 2021
|
|
2L9G
 
 | |
2LP0
 
 | |
1TKQ
 
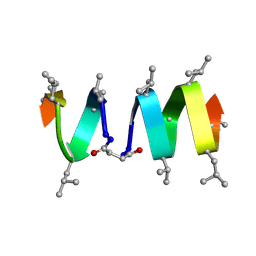 | | SOLUTION STRUCTURE OF A LINKED UNSYMMETRIC GRAMICIDIN IN A MEMBRANE-ISOELECTRICAL SOLVENTS MIXTURE IN THE PRESENCE OF CsCl | | Descriptor: | GRAMICIDIN A, MINI-GRAMICIDIN A, SUCCINIC ACID | | Authors: | Xie, X, Al-Momani, L, Bockelmann, D, Griesinger, C, Koert, U. | | Deposit date: | 2004-06-09 | | Release date: | 2004-07-13 | | Last modified: | 2019-09-25 | | Method: | SOLUTION NMR | | Cite: | An Asymmetric Ion Channel Derived from Gramicidin A. Synthesis, Function and NMR Structure.
FEBS J., 272, 2005
|
|
1ZY6
 
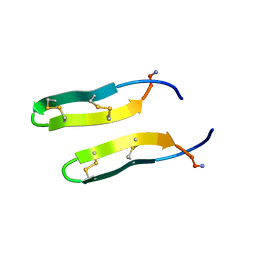 | | Membrane-bound dimer structure of Protegrin-1 (PG-1), a beta-Hairpin Antimicrobial Peptide in Lipid Bilayers from Rotational-Echo Double-Resonance Solid-State NMR | | Descriptor: | Protegrin 1 | | Authors: | Wu, X, Mani, R, Tang, M, Buffy, J.J, Waring, A.J, Sherman, M.A, Hong, M. | | Deposit date: | 2005-06-09 | | Release date: | 2006-06-13 | | Last modified: | 2022-03-02 | | Method: | SOLID-STATE NMR | | Cite: | Membrane-Bound Dimer Structure of a beta-Hairpin Antimicrobial Peptide from Rotational-Echo Double-Resonance Solid-State NMR.
Biochemistry, 45, 2006
|
|
2GB8
 
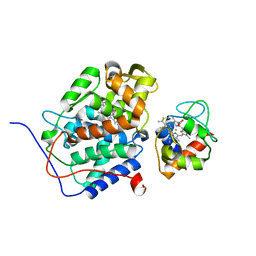 | | Solution structure of the complex between yeast iso-1-cytochrome c and yeast cytochrome c peroxidase | | Descriptor: | Cytochrome c iso-1, Cytochrome c peroxidase, HEME C, ... | | Authors: | Volkov, A.N, Worrall, J.A.R, Ubbink, M. | | Deposit date: | 2006-03-10 | | Release date: | 2006-11-21 | | Last modified: | 2022-03-09 | | Method: | SOLUTION NMR | | Cite: | Solution structure and dynamics of the complex between cytochrome c and cytochrome c peroxidase determined by paramagnetic NMR.
Proc.Natl.Acad.Sci.Usa, 103, 2006
|
|
2N9U
 
 | |
2JTI
 
 | |
6H5H
 
 | |
