6OFO
 
 | | Crystal structure of split green fluorescent protein (GFP); s10 circular permutant (194-195) | | Descriptor: | Green fluorescent protein (GFP); s10 circular permutant (194-195) | | Authors: | Lin, C.-Y, Romei, M.G, Deller, M.C, Doukov, T.I, Boxer, S.G. | | Deposit date: | 2019-03-31 | | Release date: | 2019-07-10 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.603 Å) | | Cite: | Unified Model for Photophysical and Electro-Optical Properties of Green Fluorescent Proteins.
J.Am.Chem.Soc., 141, 2019
|
|
1GGX
 
 | |
6MWQ
 
 | |
6FWW
 
 | | GFP/KKK. A redesigned GFP with improved solubility | | Descriptor: | Green fluorescent protein | | Authors: | Varejao, N, Lascorz, J, Gil-Garcia, M, Diaz-Caballero, M, Navarro, S, Ventura, S, Reverter, D. | | Deposit date: | 2018-03-07 | | Release date: | 2018-08-01 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.131 Å) | | Cite: | Combining Structural Aggregation Propensity and Stability Predictions To Redesign Protein Solubility.
Mol. Pharm., 15, 2018
|
|
3MGF
 
 | | Crystal Structure of Monomeric Kusabira-Orange (MKO), Orange-Emitting GFP-like Protein, at pH 7.5 | | Descriptor: | Fluorescent protein | | Authors: | Ebisawa, T, Yamamura, A, Ohtsuka, J, Kameda, Y, Hayakawa, K, Nagata, K, Tanokura, M. | | Deposit date: | 2010-04-06 | | Release date: | 2011-03-16 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal Structure of Monomeric Kusabira-Orange (MKO), Orange-Emitting GFP-like Protein, at pH 7.5
To be Published
|
|
4KF5
 
 | | Crystal Structure of Split GFP complexed with engineered sfCherry with an insertion of GFP fragment | | Descriptor: | fluorescent protein GFP1-9, fluorescent protein sfCherry+GFP10-11 | | Authors: | Nguyen, H.B, Hung, L.-W, Yeates, T.O, Waldo, G.S, Terwilliger, T.C. | | Deposit date: | 2013-04-26 | | Release date: | 2013-12-18 | | Method: | X-RAY DIFFRACTION (2.599 Å) | | Cite: | Split green fluorescent protein as a modular binding partner for protein crystallization.
Acta Crystallogr.,Sect.D, 69, 2013
|
|
4GOB
 
 | | Low pH Crystal Structure of a reconstructed Kaede-type Red Fluorescent Protein, Least Evolved Ancestor (LEA) | | Descriptor: | Kaede-type Fluorescent Protein | | Authors: | Kim, H, Grunkemeyer, T.J, Chen, L, Fromme, R, Wachter, R.M. | | Deposit date: | 2012-08-19 | | Release date: | 2013-07-31 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.53 Å) | | Cite: | Acid-base catalysis and crystal structures of a least evolved ancestral GFP-like protein undergoing green-to-red photoconversion.
Biochemistry, 52, 2013
|
|
6RUM
 
 | | Crystal structure of GFP-LAMA-G97 - a GFP enhancer nanobody with cpDHFR insertion and TMP and NADPH | | Descriptor: | CHLORIDE ION, DI(HYDROXYETHYL)ETHER, GFP-LAMA-G97 a GFP enhancer nanobody with cpDHFR insertion, ... | | Authors: | Farrants, H, Tarnawski, M, Mueller, T.G, Otsuka, S, Hiblot, J, Koch, B, Kueblbeck, M, Kraeusslich, H.-G, Ellenberg, J, Johnsson, K. | | Deposit date: | 2019-05-28 | | Release date: | 2020-02-12 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Chemogenetic Control of Nanobodies.
Nat.Methods, 17, 2020
|
|
1RRX
 
 | | Crystallographic Evidence for Isomeric Chromophores in 3-Fluorotyrosyl-Green Fluorescent Protein | | Descriptor: | SIGF1-GFP fusion protein | | Authors: | Bae, J.H, Paramita Pal, P, Moroder, L, Huber, R, Budisa, N. | | Deposit date: | 2003-12-09 | | Release date: | 2004-06-08 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystallographic Evidence for Isomeric Chromophores in 3-Fluorotyrosyl-Green Fluorescent Protein.
Chembiochem, 5, 2004
|
|
4PPJ
 
 | | Crystal structure of Phanta, a weakly fluorescent photochromic GFP-like protein. ON state | | Descriptor: | Monomeric Azami Green | | Authors: | Don Paul, C, Traore, D.A.K, Devenish, R.J, Close, D, Bell, T, Bradbury, A, Wilce, M.C.J, Prescott, M. | | Deposit date: | 2014-02-27 | | Release date: | 2015-04-08 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | X-Ray Crystal Structure and Properties of Phanta, a Weakly Fluorescent Photochromic GFP-Like Protein.
Plos One, 10, 2015
|
|
3KPX
 
 | | Crystal Structure Analysis of photoprotein clytin | | Descriptor: | Apophotoprotein clytin-3, C2-HYDROPEROXY-COELENTERAZINE, CALCIUM ION | | Authors: | Titushin, M.S, Li, Y, Stepanyuk, G.A, Wang, B.-C, Lee, J, Vysotski, E.S, Liu, Z.-J. | | Deposit date: | 2009-11-17 | | Release date: | 2010-10-06 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.899 Å) | | Cite: | NMR derived topology of a GFP-photoprotein energy transfer complex
J.Biol.Chem., 285, 2010
|
|
1Z1P
 
 | |
7RRH
 
 | | Crystal structure of fast switching R66M/M159T mutant of fluorescent protein Dronpa (Dronpa2) | | Descriptor: | Fluorescent protein Dronpa | | Authors: | Lin, C.-Y, Romei, M.G, Mathews, I.I, Boxer, S.G. | | Deposit date: | 2021-08-09 | | Release date: | 2021-10-13 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.747 Å) | | Cite: | Energetic Basis and Design of Enzyme Function Demonstrated Using GFP, an Excited-State Enzyme.
J.Am.Chem.Soc., 144, 2022
|
|
7RRK
 
 | | Crystal structure of fast switching M159E mutant of fluorescent protein Dronpa (Dronpa2) | | Descriptor: | Fluorescent protein Dronpa | | Authors: | Lin, C.-Y, Romei, M.G, Mathews, I.I, Boxer, S.G. | | Deposit date: | 2021-08-09 | | Release date: | 2021-10-13 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.929 Å) | | Cite: | Energetic Basis and Design of Enzyme Function Demonstrated Using GFP, an Excited-State Enzyme.
J.Am.Chem.Soc., 144, 2022
|
|
7RRJ
 
 | | Crystal structure of fast switching M159Q mutant of fluorescent protein Dronpa (Dronpa2) | | Descriptor: | Fluorescent protein Dronpa | | Authors: | Lin, C.-Y, Romei, M.G, Mathews, I.I, Boxer, S.G. | | Deposit date: | 2021-08-09 | | Release date: | 2021-10-13 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Energetic Basis and Design of Enzyme Function Demonstrated Using GFP, an Excited-State Enzyme.
J.Am.Chem.Soc., 144, 2022
|
|
7RRI
 
 | | Crystal structure of fast switching S142A/M159T mutant of fluorescent protein Dronpa (Dronpa2) | | Descriptor: | Fluorescent protein Dronpa | | Authors: | Lin, C.-Y, Romei, M.G, Mathews, I.I, Boxer, S.G. | | Deposit date: | 2021-08-09 | | Release date: | 2021-10-13 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.643 Å) | | Cite: | Energetic Basis and Design of Enzyme Function Demonstrated Using GFP, an Excited-State Enzyme.
J.Am.Chem.Soc., 144, 2022
|
|
4Q9R
 
 | | Crystal structure of an RNA aptamer bound to trifluoroethyl-ligand analog in complex with Fab | | Descriptor: | (5Z)-5-(3,5-difluoro-4-hydroxybenzylidene)-2-methyl-3-(2,2,2-trifluoroethyl)-3,5-dihydro-4H-imidazol-4-one, Fab BL3-6, HEAVY CHAIN, ... | | Authors: | Huang, H, Suslov, N.B, Li, N.-S, Shelke, S.A, Evans, M.E, Koldobskaya, Y, Rice, P.A, Piccirilli, J.A. | | Deposit date: | 2014-05-01 | | Release date: | 2014-06-18 | | Last modified: | 2017-07-26 | | Method: | X-RAY DIFFRACTION (3.12 Å) | | Cite: | A G-quadruplex-containing RNA activates fluorescence in a GFP-like fluorophore.
Nat.Chem.Biol., 10, 2014
|
|
4Q9Q
 
 | | Crystal structure of an RNA aptamer bound to bromo-ligand analog in complex with Fab | | Descriptor: | (5Z)-5-(3-bromobenzylidene)-2,3-dimethyl-3,5-dihydro-4H-imidazol-4-one, Fab BL3-6, HEAVY CHAIN, ... | | Authors: | Huang, H, Suslov, N.B, Li, N.-S, Shelke, S.A, Evans, M.E, Koldobskaya, Y, Rice, P.A, Piccirilli, J.A. | | Deposit date: | 2014-05-01 | | Release date: | 2014-06-18 | | Last modified: | 2017-07-26 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | A G-quadruplex-containing RNA activates fluorescence in a GFP-like fluorophore.
Nat.Chem.Biol., 10, 2014
|
|
8AM4
 
 | | Cl-rsEGFP2 Long Wavelength Structure | | Descriptor: | Green fluorescent protein | | Authors: | Orr, C.M, Fadini, A, van Thor, J. | | Deposit date: | 2022-08-02 | | Release date: | 2023-08-02 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.02 Å) | | Cite: | Serial Femtosecond Crystallography Reveals that Photoactivation in a Fluorescent Protein Proceeds via the Hula Twist Mechanism.
J.Am.Chem.Soc., 2023
|
|
8FI8
 
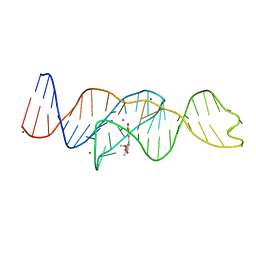 | |
8FHV
 
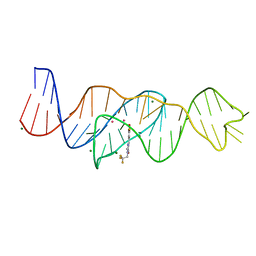 | | Structure of Lettuce aptamer bound to DFHBI-1T with thallium I ions | | Descriptor: | (5Z)-5-(3,5-difluoro-4-hydroxybenzylidene)-2-methyl-3-(2,2,2-trifluoroethyl)-3,5-dihydro-4H-imidazol-4-one, Lettuce DNA aptamer, MAGNESIUM ION, ... | | Authors: | Passalacqua, L.F.M, Ferre-D'Amare, A.R. | | Deposit date: | 2022-12-15 | | Release date: | 2023-05-10 | | Last modified: | 2023-07-05 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Intricate 3D architecture of a DNA mimic of GFP.
Nature, 618, 2023
|
|
8FHZ
 
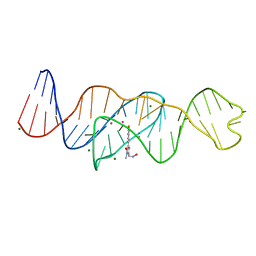 | | Structure of Lettuce aptamer bound to DFHO | | Descriptor: | (5Z)-5-[(3,5-difluoro-4-hydroxyphenyl)methylidene]-2-[(E)-(hydroxyimino)methyl]-3-methyl-3,5-dihydro-4H-imidazol-4-one, Lettuce DNA aptamer, MAGNESIUM ION, ... | | Authors: | Passalacqua, L.F.M, Ferre-D'Amare, A.R. | | Deposit date: | 2022-12-15 | | Release date: | 2023-05-10 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Intricate 3D architecture of a DNA mimic of GFP.
Nature, 618, 2023
|
|
8FI2
 
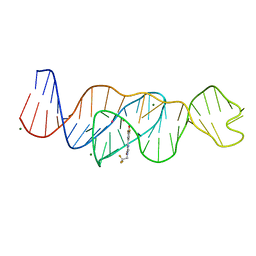 | | Structure of Lettuce C20T bound to DFHBI-1T | | Descriptor: | (5Z)-5-(3,5-difluoro-4-hydroxybenzylidene)-2-methyl-3-(2,2,2-trifluoroethyl)-3,5-dihydro-4H-imidazol-4-one, Lettuce DNA aptamer, MAGNESIUM ION, ... | | Authors: | Passalacqua, L.F.M, Ferre-D'Amare, A.R. | | Deposit date: | 2022-12-15 | | Release date: | 2023-05-10 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Intricate 3D architecture of a DNA mimic of GFP.
Nature, 618, 2023
|
|
8FI1
 
 | | Structure of Lettuce C20G bound to DFHO | | Descriptor: | (5Z)-5-[(3,5-difluoro-4-hydroxyphenyl)methylidene]-2-[(E)-(hydroxyimino)methyl]-3-methyl-3,5-dihydro-4H-imidazol-4-one, Lettuce DNA aptamer, MAGNESIUM ION, ... | | Authors: | Passalacqua, L.F.M, Ferre-D'Amare, A.R. | | Deposit date: | 2022-12-15 | | Release date: | 2023-05-10 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Intricate 3D architecture of a DNA mimic of GFP.
Nature, 618, 2023
|
|
8FI0
 
 | |
