2XU3
 
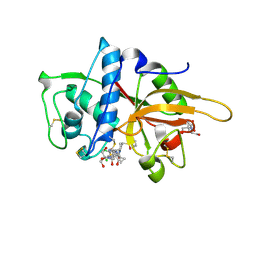 | | CATHEPSIN L WITH A NITRILE INHIBITOR | | 分子名称: | (2S,4R)-4-(2-chlorophenyl)sulfonyl-1-[1-(5-chlorothiophen-2-yl)cyclopropyl]carbonyl-N-[1-(iminomethyl)cyclopropyl]pyrrolidine-2-carboxamide, 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CATHEPSIN L1, ... | | 著者 | Banner, D.W, Benz, J.M, Haap, W. | | 登録日 | 2010-10-14 | | 公開日 | 2011-01-12 | | 最終更新日 | 2023-12-20 | | 実験手法 | X-RAY DIFFRACTION (0.9 Å) | | 主引用文献 | Systematic Investigation of Halogen Bonding in Protein-Ligand Interactions.
Angew.Chem.Int.Ed.Engl., 50, 2011
|
|
2BW4
 
 | | Atomic Resolution Structure of Resting State of the Achromobacter cycloclastes Cu Nitrite Reductase | | 分子名称: | ACETATE ION, COPPER (II) ION, COPPER-CONTAINING NITRITE REDUCTASE, ... | | 著者 | Antonyuk, S.V, Strange, R.W, Sawers, G, Eady, R.R, Hasnain, S.S. | | 登録日 | 2005-07-12 | | 公開日 | 2005-08-17 | | 最終更新日 | 2023-12-13 | | 実験手法 | X-RAY DIFFRACTION (0.9 Å) | | 主引用文献 | Atomic Resolution Structures of Resting-State, Substrate- and Product-Complexed Cu-Nitrite Reductase Provide Insight Into Catalytic Mechanism
Proc.Natl.Acad.Sci.USA, 102, 2005
|
|
4NPD
 
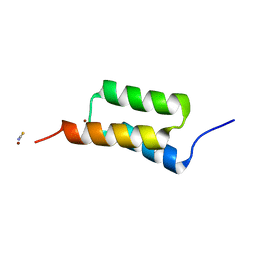 | | High-resolution structure of C domain of staphylococcal protein A at cryogenic temperature | | 分子名称: | Immunoglobulin G-binding protein A, THIOCYANATE ION, ZINC ION | | 著者 | Deis, L.N, Pemble IV, C.W, Oas, T.G, Richardson, J.S, Richardson, D.C. | | 登録日 | 2013-11-21 | | 公開日 | 2014-10-08 | | 最終更新日 | 2024-02-28 | | 実験手法 | X-RAY DIFFRACTION (0.9 Å) | | 主引用文献 | Multiscale conformational heterogeneity in staphylococcal protein a: possible determinant of functional plasticity.
Structure, 22, 2014
|
|
2YL7
 
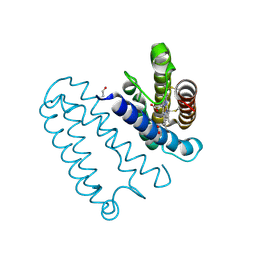 | | CYTOCHROME C PRIME FROM ALCALIGENES XYLOSOXIDANS: AS ISOLATED L16G VARIANT AT 0.9 A RESOLUTION - RESTRAINT REFINEMENT | | 分子名称: | CARBON MONOXIDE, CYTOCHROME C', HEME C | | 著者 | Antonyuk, S.V, Rustage, N, Eady, R.R, Hasnain, S.S. | | 登録日 | 2011-05-31 | | 公開日 | 2011-10-05 | | 最終更新日 | 2023-12-20 | | 実験手法 | X-RAY DIFFRACTION (0.9 Å) | | 主引用文献 | Carbon Monoxide Poisoning is Prevented by the Energy Costs of Conformational Changes in Gas- Binding Haemproteins.
Proc.Natl.Acad.Sci.USA, 108, 2011
|
|
6XVM
 
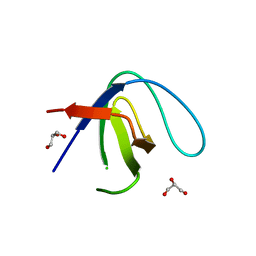 | |
2PEV
 
 | | Complex of Aldose Reductase with NADP+ and simaltaneously bound competetive inhibitors Fidarestat and IDD594. Concentration of Fidarestat in soaking solution exceeds concentration of IDD594. | | 分子名称: | (2S,4S)-2-AMINOFORMYL-6-FLUORO-SPIRO[CHROMAN-4,4'-IMIDAZOLIDINE]-2',5'-DIONE, Aldose reductase, CHLORIDE ION, ... | | 著者 | Petrova, T, Hazemann, I, Cousido, A, Mitschler, A, Ginell, S, Joachimiak, A, Podjarny, A. | | 登録日 | 2007-04-03 | | 公開日 | 2007-04-17 | | 最終更新日 | 2023-08-30 | | 実験手法 | X-RAY DIFFRACTION (0.9 Å) | | 主引用文献 | Crystal packing modifies ligand binding affinity: The case of aldose reductase.
Proteins, 80, 2012
|
|
3K34
 
 | | Human carbonic anhydrase II with a sulfonamide inhibitor | | 分子名称: | (4-SULFAMOYL-PHENYL)-THIOCARBAMIC ACID O-(2-THIOPHEN-3-YL-ETHYL) ESTER, 4-(HYDROXYMERCURY)BENZOIC ACID, Carbonic anhydrase 2, ... | | 著者 | Behnke, C.A, Le Trong, I, Merritt, E.A, Teller, D.C, Stenkamp, R.E. | | 登録日 | 2009-10-01 | | 公開日 | 2010-05-12 | | 最終更新日 | 2023-09-06 | | 実験手法 | X-RAY DIFFRACTION (0.9 Å) | | 主引用文献 | Atomic resolution studies of carbonic anhydrase II.
Acta Crystallogr.,Sect.D, 66, 2010
|
|
1VYR
 
 | | Structure of pentaerythritol tetranitrate reductase complexed with picric acid | | 分子名称: | FLAVIN MONONUCLEOTIDE, PENTAERYTHRITOL TETRANITRATE REDUCTASE, PICRIC ACID | | 著者 | Barna, T, Moody, P.C.E. | | 登録日 | 2004-05-05 | | 公開日 | 2004-06-10 | | 最終更新日 | 2023-12-13 | | 実験手法 | X-RAY DIFFRACTION (0.9 Å) | | 主引用文献 | Atomic Resolution Structures and Solution Behavior of Enzyme-Substrate Complexes of Enterobacter Cloacae Pb2 Pentaerythritol Tetranitrate Reductase: Multiple Conformational States and Implications for the Mechanism of Nitroaromatic Explosive Degradation
J.Biol.Chem., 279, 2004
|
|
4NSV
 
 | | Lysobacter enzymogenes lysc endoproteinase K30R mutant covalently inhibited by TLCK | | 分子名称: | CHLORIDE ION, Lysyl endopeptidase, N-[(2S,3S)-7-amino-1-chloro-2-hydroxyheptan-3-yl]-4-methylbenzenesulfonamide (Bound Form), ... | | 著者 | Asztalos, P, Muller, A, Holke, W, Sobek, H, Rudolph, M.G. | | 登録日 | 2013-11-29 | | 公開日 | 2014-04-23 | | 最終更新日 | 2023-09-20 | | 実験手法 | X-RAY DIFFRACTION (0.9 Å) | | 主引用文献 | Atomic resolution structure of a lysine-specific endoproteinase from Lysobacter enzymogenes suggests a hydroxyl group bound to the oxyanion hole.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
2IDQ
 
 | | Structure of M98A mutant of amicyanin, Cu(II) | | 分子名称: | Amicyanin, COPPER (II) ION, PHOSPHATE ION | | 著者 | Carrell, C.J, Ma, J.K, Antholine, W, Hosler, J.P, Mathews, F.S, Davidson, V.L. | | 登録日 | 2006-09-15 | | 公開日 | 2007-03-13 | | 最終更新日 | 2023-08-30 | | 実験手法 | X-RAY DIFFRACTION (0.9 Å) | | 主引用文献 | Generation of Novel Copper Sites by Mutation of the Axial Ligand of Amicyanin. Atomic Resolution Structures and Spectroscopic Properties
Biochemistry, 46, 2007
|
|
6FMC
 
 | | Neuropilin1-b1 domain in complex with EG01377, 0.9 Angstrom structure | | 分子名称: | (2~{S})-2-[[3-[[5-[4-(aminomethyl)phenyl]-1-benzofuran-7-yl]sulfonylamino]thiophen-2-yl]carbonylamino]-5-carbamimidamido-pentanoic acid, Neuropilin-1 | | 著者 | Yelland, T, Djordjevic, S, Fotinou, K, Selwood, D, Zachary, I, Frankel, P. | | 登録日 | 2018-01-30 | | 公開日 | 2018-10-17 | | 最終更新日 | 2024-01-17 | | 実験手法 | X-RAY DIFFRACTION (0.9 Å) | | 主引用文献 | Small Molecule Neuropilin-1 Antagonists Combine Antiangiogenic and Antitumor Activity with Immune Modulation through Reduction of Transforming Growth Factor Beta (TGF beta ) Production in Regulatory T-Cells.
J. Med. Chem., 61, 2018
|
|
6B00
 
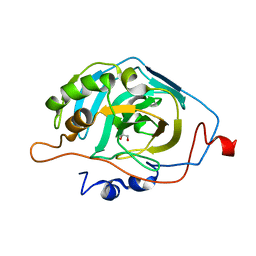 | |
5R32
 
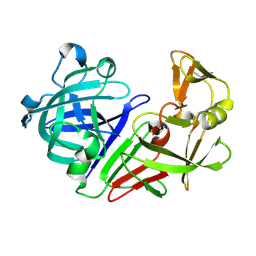 | | PanDDA analysis group deposition -- Auto-refined data of Endothiapepsin for ground state model 26, DMSO-Free | | 分子名称: | Endothiapepsin | | 著者 | Wollenhaupt, J, Metz, A, Barthel, T, Lima, G.M.A, Heine, A, Mueller, U, Klebe, G, Weiss, M.S. | | 登録日 | 2020-02-13 | | 公開日 | 2020-06-03 | | 最終更新日 | 2020-07-08 | | 実験手法 | X-RAY DIFFRACTION (0.9 Å) | | 主引用文献 | F2X-Universal and F2X-Entry: Structurally Diverse Compound Libraries for Crystallographic Fragment Screening.
Structure, 28, 2020
|
|
6LK1
 
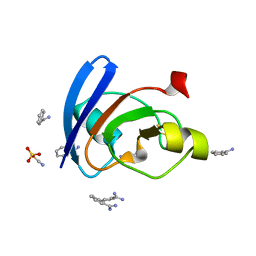 | | Ultrahigh resolution X-ray structure of Ferredoxin I from C. reinhardtii | | 分子名称: | BENZAMIDINE, FE2/S2 (INORGANIC) CLUSTER, Ferredoxin, ... | | 著者 | Onishi, Y, Kurisu, G, Tanaka, H. | | 登録日 | 2019-12-17 | | 公開日 | 2020-05-27 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (0.9 Å) | | 主引用文献 | X-ray dose-dependent structural changes of the [2Fe-2S] ferredoxin from Chlamydomonas reinhardtii.
J.Biochem., 167, 2020
|
|
1G66
 
 | | ACETYLXYLAN ESTERASE AT 0.90 ANGSTROM RESOLUTION | | 分子名称: | ACETYL XYLAN ESTERASE II, GLYCEROL, SULFATE ION | | 著者 | Ghosh, D, Sawicki, M, Lala, P, Erman, M, Pangborn, W, Eyzaguirre, J, Gutierrez, R, Jornvall, H, Thiel, D.J. | | 登録日 | 2000-11-03 | | 公開日 | 2001-01-17 | | 最終更新日 | 2011-07-13 | | 実験手法 | X-RAY DIFFRACTION (0.9 Å) | | 主引用文献 | Multiple conformations of catalytic serine and histidine in acetylxylan esterase at 0.90 A.
J.Biol.Chem., 276, 2001
|
|
5RBR
 
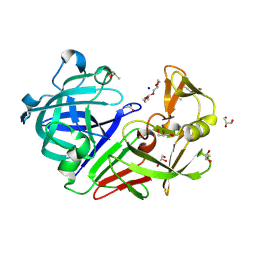 | | PanDDA analysis group deposition -- Endothiapepsin changed state model for fragment F2X-Entry Library C04a | | 分子名称: | 2-hydrazinyl-4-methoxypyrimidine, ACETATE ION, DIMETHYL SULFOXIDE, ... | | 著者 | Weiss, M.S, Wollenhaupt, J, Metz, A, Barthel, T, Lima, G.M.A, Heine, A, Mueller, U, Klebe, G. | | 登録日 | 2020-03-24 | | 公開日 | 2020-06-03 | | 最終更新日 | 2024-04-24 | | 実験手法 | X-RAY DIFFRACTION (0.9 Å) | | 主引用文献 | F2X-Universal and F2X-Entry: Structurally Diverse Compound Libraries for Crystallographic Fragment Screening.
Structure, 28, 2020
|
|
6M9I
 
 | | L-GSTSTA from degenerate octameric repeats in InaZ, residues 707-712 | | 分子名称: | Ice nucleation protein | | 著者 | Zee, C, Glynn, C, Gallagher-Jones, M, Miao, J, Santiago, C.G, Cascio, D, Gonen, T, Sawaya, M.R, Rodriguez, J.A. | | 登録日 | 2018-08-23 | | 公開日 | 2019-03-27 | | 最終更新日 | 2024-03-13 | | 実験手法 | ELECTRON CRYSTALLOGRAPHY (0.9 Å) | | 主引用文献 | Homochiral and racemic MicroED structures of a peptide repeat from the ice-nucleation protein InaZ.
IUCrJ, 6, 2019
|
|
3VOR
 
 | | Crystal Structure Analysis of the CofA | | 分子名称: | CFA/III pilin | | 著者 | Fukakusa, S, Kawahara, K, Nakamura, S, Iwasita, T, Baba, S, Nishimura, M, Kobayashi, Y, Honda, T, Iida, T, Taniguchi, T, Ohkubo, T. | | 登録日 | 2012-02-06 | | 公開日 | 2012-09-26 | | 最終更新日 | 2013-07-31 | | 実験手法 | X-RAY DIFFRACTION (0.9 Å) | | 主引用文献 | Structure of the CFA/III major pilin subunit CofA from human enterotoxigenic Escherichia coli determined at 0.90 A resolution by sulfur-SAD phasing
Acta Crystallogr.,Sect.D, 68, 2012
|
|
6BZM
 
 | | GFGNFGTS from low-complexity/FG repeat domain of Nup98, residues 116-123 | | 分子名称: | Nuclear pore complex protein Nup98-Nup96 | | 著者 | Hughes, M.P, Rodriguez, J.A, Sawaya, M.R, Cascio, D, Chong, L, Gonen, T, Eisenberg, D.S. | | 登録日 | 2017-12-24 | | 公開日 | 2018-04-04 | | 最終更新日 | 2024-03-13 | | 実験手法 | ELECTRON CRYSTALLOGRAPHY (0.9 Å) | | 主引用文献 | Atomic structures of low-complexity protein segments reveal kinked beta sheets that assemble networks.
Science, 359, 2018
|
|
6M9J
 
 | | Racemic-GSTSTA from degenerate octameric repeats in InaZ, residues 707-712 | | 分子名称: | Ice nucleation protein | | 著者 | Zee, C, Glynn, C, Gallagher-Jones, M, Miao, J, Santiago, C.G, Cascio, D, Gonen, T, Sawaya, M.R, Rodriguez, J.A. | | 登録日 | 2018-08-23 | | 公開日 | 2019-03-27 | | 最終更新日 | 2024-03-13 | | 実験手法 | ELECTRON CRYSTALLOGRAPHY (0.9 Å) | | 主引用文献 | Homochiral and racemic MicroED structures of a peptide repeat from the ice-nucleation protein InaZ.
IUCrJ, 6, 2019
|
|
2DCG
 
 | | MOLECULAR STRUCTURE OF A LEFT-HANDED DOUBLE HELICAL DNA FRAGMENT AT ATOMIC RESOLUTION | | 分子名称: | DNA (5'-D(*CP*GP*CP*GP*CP*G)-3'), MAGNESIUM ION, SPERMINE | | 著者 | Wang, A.H.-J, Quigley, G.J, Kolpak, F.J, Crawford, J.L, Van Boom, J.H, Van Der Marel, G.A, Rich, A. | | 登録日 | 1988-08-29 | | 公開日 | 1989-01-09 | | 最終更新日 | 2024-02-14 | | 実験手法 | X-RAY DIFFRACTION (0.9 Å) | | 主引用文献 | Molecular structure of a left-handed double helical DNA fragment at atomic resolution.
Nature, 282, 1979
|
|
3KS3
 
 | |
4EA9
 
 | | X-ray structure of GDP-perosamine N-acetyltransferase in complex with transition state analog at 0.9 Angstrom resolution | | 分子名称: | CHLORIDE ION, GDP-N-acetylperosamine-coenzyme A, Perosamine N-acetyltransferase | | 著者 | Thoden, J.B, Reinhardt, L.A, Cook, P.D, Menden, P, Cleland, W.W, Holden, H.M. | | 登録日 | 2012-03-22 | | 公開日 | 2012-04-04 | | 最終更新日 | 2023-09-13 | | 実験手法 | X-RAY DIFFRACTION (0.9 Å) | | 主引用文献 | Catalytic Mechanism of Perosamine N-Acetyltransferase Revealed by High-Resolution X-ray Crystallographic Studies and Kinetic Analyses.
Biochemistry, 51, 2012
|
|
1M24
 
 | |
3ZR8
 
 | | Crystal structure of RxLR effector Avr3a11 from Phytophthora capsici | | 分子名称: | AVR3A11, CHLORIDE ION, TRIETHYLENE GLYCOL | | 著者 | Boutemy, L.S, King, S.R.F, Win, J, Hughes, R.K, Clarke, T.A, Blumenschein, T.M.A, Kamoun, S, Banfield, M.J. | | 登録日 | 2011-06-15 | | 公開日 | 2011-08-03 | | 最終更新日 | 2011-10-19 | | 実験手法 | X-RAY DIFFRACTION (0.9 Å) | | 主引用文献 | Structures of Phytophthora Rxlr Effector Proteins: A Conserved But Adaptable Fold Underpins Functional Diversity.
J.Biol.Chem., 286, 2011
|
|
