6UGC
 
 | | C3 symmetric peptide design number 3 | | 分子名称: | C3-3 cyclic peptide design, CADMIUM ION, SODIUM ION | | 著者 | Mulligan, V.K, Kang, C.S, Antselovich, I, Sawaya, M.R, Yeates, T.O, Baker, D. | | 登録日 | 2019-09-26 | | 公開日 | 2020-12-02 | | 実験手法 | X-RAY DIFFRACTION (0.9 Å) | | 主引用文献 | Computational design of mixed chirality peptide macrocycles with internal symmetry.
Protein Sci., 29, 2020
|
|
7PSY
 
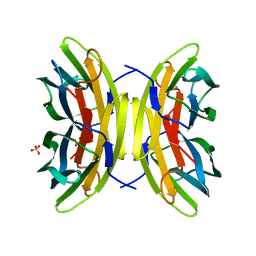 | | X-ray crystal structure of perdeuterated LecB lectin in complex with perdeuterated fucose | | 分子名称: | CALCIUM ION, Fucose-binding lectin, SULFATE ION, ... | | 著者 | Gajdos, L, Blakeley, M.P, Haertlein, M, Forsyth, T.V, Devos, J.M, Imberty, A. | | 登録日 | 2021-09-24 | | 公開日 | 2022-01-12 | | 最終更新日 | 2024-01-31 | | 実験手法 | X-RAY DIFFRACTION (0.9 Å) | | 主引用文献 | Neutron crystallography reveals mechanisms used by Pseudomonas aeruginosa for host-cell binding.
Nat Commun, 13, 2022
|
|
6UOS
 
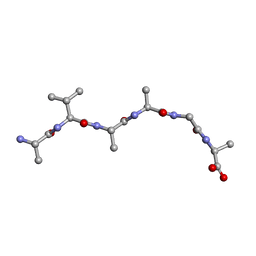 | |
6UOR
 
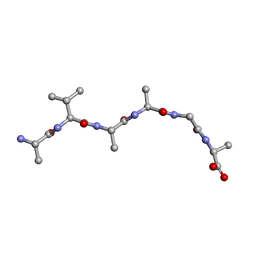 | |
7WDJ
 
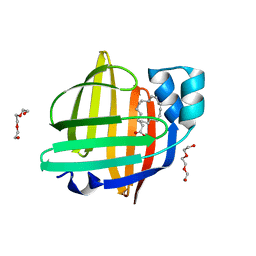 | | The 0.90 angstrom X-ray structure of the human heart fatty acid-binding protein complexed with linoleic acid | | 分子名称: | Fatty acid-binding protein, heart, HEXAETHYLENE GLYCOL, ... | | 著者 | Sugiyama, S, Matsuoka, S, Tsuchikawa, H, Sonoyama, M, Inoue, Y, Hayashi, F, Murata, M. | | 登録日 | 2021-12-21 | | 公開日 | 2022-12-21 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (0.9 Å) | | 主引用文献 | The 0.90 angstrom X-ray structure of the human heart fatty acid-binding protein complexed with linoleic acid
To Be Published
|
|
7WPG
 
 | | The 0.90 angstrom X-ray structure of the human heart fatty acid-binding protein complexed with heptanoic acid | | 分子名称: | Fatty acid-binding protein, heart, HEXAETHYLENE GLYCOL, ... | | 著者 | Sugiyama, S, Matsuoka, S, Tsuchikawa, H, Sonoyama, M, Inoue, Y, Hayashi, F, Murata, M. | | 登録日 | 2022-01-23 | | 公開日 | 2023-01-25 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (0.9 Å) | | 主引用文献 | The 0.90 angstrom X-ray structure of the human heart fatty acid-binding protein complexed with heptanoic acid
To Be Published
|
|
7WOM
 
 | | The 0.90 angstrom X-ray structure of the human heart fatty acid-binding protein complexed with eicosapentaenoic acid | | 分子名称: | 5,8,11,14,17-EICOSAPENTAENOIC ACID, Fatty acid-binding protein, heart | | 著者 | Sugiyama, S, Kakinouchi, K, Nakano, R, Matsuoka, S, Tsuchikawa, H, Sonoyama, M, Inoue, Y, Hayashi, F, Murata, M. | | 登録日 | 2022-01-21 | | 公開日 | 2023-01-25 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (0.9 Å) | | 主引用文献 | The 0.90 angstrom X-ray structure of the human heart fatty acid-binding protein complexed with eicosapentaenoic acid
To Be Published
|
|
6DIY
 
 | | YTFGQ segment from Human Immunoglobulin Light-Chain Variable Domain, Residues 96-100, assembled as an amyloid fibril | | 分子名称: | YTFGQ segment Light-Chain Variable Domain Kappa AL09 | | 著者 | Brumshtein, B, Esswein, S.R, Sawaya, M.R, Eisenberg, D.S. | | 登録日 | 2018-05-24 | | 公開日 | 2018-10-31 | | 最終更新日 | 2024-03-13 | | 実験手法 | X-RAY DIFFRACTION (0.9 Å) | | 主引用文献 | Identification of two principal amyloid-driving segments in variable domains of Ig light chains in systemic light-chain amyloidosis.
J. Biol. Chem., 293, 2018
|
|
7N2L
 
 | | MicroED structure of a mutant mammalian prion segment | | 分子名称: | prion protein | | 著者 | Flores, M.D, Richards, L.S, Zee, C.T, Glynn, C, Gallagher-Jones, M, Sawaya, M.R. | | 登録日 | 2021-05-29 | | 公開日 | 2022-06-01 | | 最終更新日 | 2023-05-10 | | 実験手法 | ELECTRON CRYSTALLOGRAPHY (0.9 Å) | | 主引用文献 | Fragment-Based Ab Initio Phasing of Peptidic Nanocrystals by MicroED.
Acs Bio Med Chem Au, 3, 2023
|
|
3SBN
 
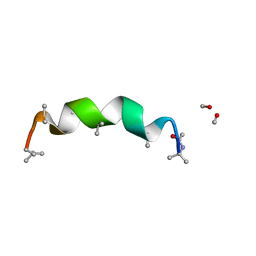 | | trichovirin I-4A in polar environment at 0.9 Angstroem | | 分子名称: | ACETONITRILE, METHANOL, Trichovirin I-4A | | 著者 | Gessmann, R, Axford, D, Petratos, K. | | 登録日 | 2011-06-06 | | 公開日 | 2011-12-28 | | 最終更新日 | 2023-11-15 | | 実験手法 | X-RAY DIFFRACTION (0.9 Å) | | 主引用文献 | Four complete turns of a curved 310-helix at atomic resolution: The crystal structure of the peptaibol trichovirin I-4A in polar environment suggests a transition to alpha-helix for membrane function
Acta Crystallogr.,Sect.D, 68, 2012
|
|
5Y2S
 
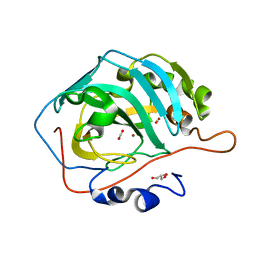 | |
6Y14
 
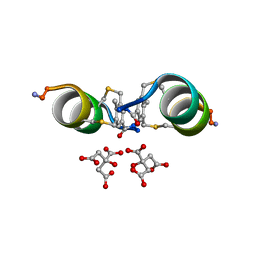 | |
7QP2
 
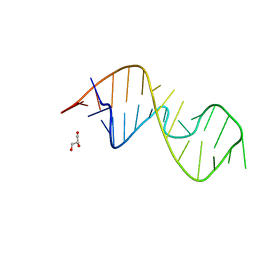 | | 1-deazaguanosine modified-RNA Sarcin Ricin Loop | | 分子名称: | GLYCEROL, RNA (27-MER) | | 著者 | Ennifar, E, Micura, R, Bereiter, R, Renard, E, Kreutz, C. | | 登録日 | 2021-12-30 | | 公開日 | 2022-07-20 | | 最終更新日 | 2024-01-31 | | 実験手法 | X-RAY DIFFRACTION (0.9 Å) | | 主引用文献 | 1-Deazaguanosine-Modified RNA: The Missing Piece for Functional RNA Atomic Mutagenesis.
J.Am.Chem.Soc., 144, 2022
|
|
1O56
 
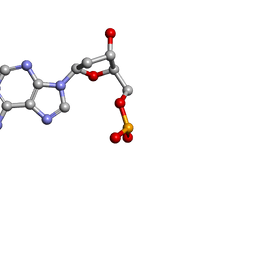 | | MOLECULAR STRUCTURE OF TWO CRYSTAL FORMS OF CYCLIC TRIADENYLIC ACID AT 1 ANGSTROM RESOLUTION | | 分子名称: | DNA (5'-CD(*AP*AP*AP*)-3') | | 著者 | Gao, Y.G, Robinson, H, Guan, Y, Liaw, Y.C, van Boom, J.H, van der Marel, G.A, Wang, A.H. | | 登録日 | 2003-08-20 | | 公開日 | 2003-08-26 | | 最終更新日 | 2023-12-27 | | 実験手法 | X-RAY DIFFRACTION (0.9 Å) | | 主引用文献 | Molecular structure of two crystal forms of cyclic triadenylic acid at 1A resolution.
J.Biomol.Struct.Dyn., 16, 1998
|
|
7TWW
 
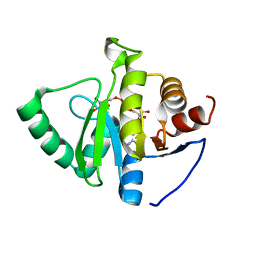 | | Crystal structure of SARS-CoV-2 NSP3 macrodomain in complex with ADP-ribose at pH 6 (P43 crystal form) | | 分子名称: | CITRIC ACID, Non-structural protein 3, [(2R,3S,4R,5R)-5-(6-AMINOPURIN-9-YL)-3,4-DIHYDROXY-OXOLAN-2-YL]METHYL [HYDROXY-[[(2R,3S,4R,5S)-3,4,5-TRIHYDROXYOXOLAN-2-YL]METHOXY]PHOSPHORYL] HYDROGEN PHOSPHATE | | 著者 | Correy, G.J, Fraser, J.S. | | 登録日 | 2022-02-07 | | 公開日 | 2022-02-23 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (0.9 Å) | | 主引用文献 | Crystal structure of SARS-CoV-2 NSP3 macrodomain in complex with ADP-ribose at pH 6 (P43 crystal form)
To Be Published
|
|
7TWN
 
 | |
7TWJ
 
 | |
7TWX
 
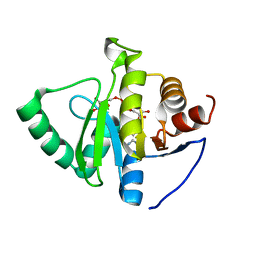 | | Crystal structure of SARS-CoV-2 NSP3 macrodomain in complex with ADP-ribose at pH 7 (P43 crystal form) | | 分子名称: | Non-structural protein 3, [(2R,3S,4R,5R)-5-(6-AMINOPURIN-9-YL)-3,4-DIHYDROXY-OXOLAN-2-YL]METHYL [HYDROXY-[[(2R,3S,4R,5S)-3,4,5-TRIHYDROXYOXOLAN-2-YL]METHOXY]PHOSPHORYL] HYDROGEN PHOSPHATE | | 著者 | Correy, G.J, Fraser, J.S. | | 登録日 | 2022-02-07 | | 公開日 | 2022-02-23 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (0.9 Å) | | 主引用文献 | Crystal structure of SARS-CoV-2 NSP3 macrodomain in complex with ADP-ribose at pH 7 (P43 crystal form)
To Be Published
|
|
7TWR
 
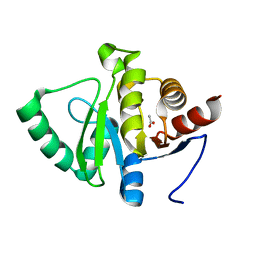 | |
7TWS
 
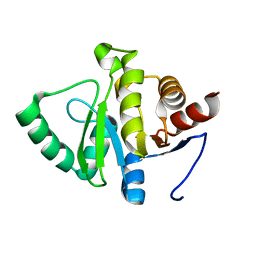 | |
7TWQ
 
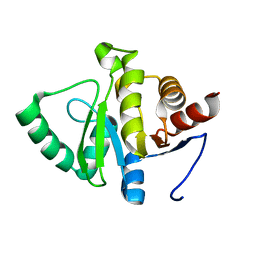 | |
7TWY
 
 | | Crystal structure of SARS-CoV-2 NSP3 macrodomain in complex with ADP-ribose at pH 8 (P43 crystal form) | | 分子名称: | Non-structural protein 3, [(2R,3S,4R,5R)-5-(6-AMINOPURIN-9-YL)-3,4-DIHYDROXY-OXOLAN-2-YL]METHYL [HYDROXY-[[(2R,3S,4R,5S)-3,4,5-TRIHYDROXYOXOLAN-2-YL]METHOXY]PHOSPHORYL] HYDROGEN PHOSPHATE | | 著者 | Correy, G.J, Fraser, J.S. | | 登録日 | 2022-02-07 | | 公開日 | 2022-02-23 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (0.9 Å) | | 主引用文献 | Crystal structure of SARS-CoV-2 NSP3 macrodomain in complex with ADP-ribose at pH 8 (P43 crystal form)
To Be Published
|
|
7TWT
 
 | | Crystal structure of SARS-CoV-2 NSP3 macrodomain in complex with ADP-ribose at pH 4 (P43 crystal form) | | 分子名称: | Non-structural protein 3, [(2R,3S,4R,5R)-5-(6-AMINOPURIN-9-YL)-3,4-DIHYDROXY-OXOLAN-2-YL]METHYL [HYDROXY-[[(2R,3S,4R,5S)-3,4,5-TRIHYDROXYOXOLAN-2-YL]METHOXY]PHOSPHORYL] HYDROGEN PHOSPHATE | | 著者 | Correy, G.J, Fraser, J.S. | | 登録日 | 2022-02-07 | | 公開日 | 2022-02-23 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (0.9 Å) | | 主引用文献 | Crystal structure of SARS-CoV-2 NSP3 macrodomain in complex with ADP-ribose at pH 4 (P43 crystal form)
To Be Published
|
|
7TX1
 
 | | Crystal structure of SARS-CoV-2 NSP3 macrodomain in complex with ADP-ribose at pH 10 (P43 crystal form) | | 分子名称: | Non-structural protein 3, [(2R,3S,4R,5R)-5-(6-AMINOPURIN-9-YL)-3,4-DIHYDROXY-OXOLAN-2-YL]METHYL [HYDROXY-[[(2R,3S,4R,5S)-3,4,5-TRIHYDROXYOXOLAN-2-YL]METHOXY]PHOSPHORYL] HYDROGEN PHOSPHATE | | 著者 | Correy, G.J, Fraser, J.S. | | 登録日 | 2022-02-07 | | 公開日 | 2022-02-23 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (0.9 Å) | | 主引用文献 | Crystal structure of SARS-CoV-2 NSP3 macrodomain in complex with ADP-ribose at pH 10 (P43 crystal form)
To Be Published
|
|
7TWO
 
 | |
