1V8Q
 
 | | Crystal structure of ribosomal protein L27 from Thermus thermophilus HB8 | | Descriptor: | 2,3-DIHYDROXY-1,4-DITHIOBUTANE, TT0826 | | Authors: | Wang, H, Takemoto-Hori, C, Murayama, K, Terada, T, Shirouzu, M, Kuramitsu, S, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2004-01-13 | | Release date: | 2004-07-13 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal structure of ribosomal protein L27 from Thermus thermophilus HB8
Protein Sci., 13, 2004
|
|
1V8R
 
 | | Crystal structure analysis of the ADP-ribose pyrophosphatase complexed with ADP-ribose and Zn | | Descriptor: | ADENOSINE-5-DIPHOSPHORIBOSE, ADP-ribose pyrophosphatase, ZINC ION | | Authors: | Yoshiba, S, Ooga, T, Nakagawa, N, Shibata, T, Inoue, Y, Yokoyama, S, Kuramitsu, S, Masui, R, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2004-01-14 | | Release date: | 2005-02-22 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural insights into the Thermus thermophilus ADP-ribose pyrophosphatase mechanism via crystal structures with the bound substrate and metal
J.Biol.Chem., 279, 2004
|
|
1V8S
 
 | | Crystal structure analusis of the ADP-ribose pyrophosphatase complexed with AMP and Mg | | Descriptor: | ADENOSINE MONOPHOSPHATE, ADP-ribose pyrophosphatase, MAGNESIUM ION | | Authors: | Yoshiba, S, Ooga, T, Nakagawa, N, Shibata, T, Inoue, Y, Yokoyama, S, Kuramitsu, S, Masui, R, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2004-01-14 | | Release date: | 2004-10-19 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural insights into the Thermus thermophilus ADP-ribose pyrophosphatase mechanism via crystal structures with the bound substrate and metal
J.Biol.Chem., 279, 2004
|
|
1V8T
 
 | | Crystal Structure analysis of the ADP-ribose pyrophosphatase complexed with ribose-5'-phosphate and Zn | | Descriptor: | ADP-ribose pyrophosphatase, RIBOSE-5-PHOSPHATE, ZINC ION | | Authors: | Yoshiba, S, Ooga, T, Nakagawa, N, Shibata, T, Inoue, Y, Yokoyama, S, Kuramitsu, S, Masui, R, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2004-01-14 | | Release date: | 2004-10-19 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural insights into the Thermus thermophilus ADP-ribose pyrophosphatase mechanism via crystal structures with the bound substrate and metal
J.Biol.Chem., 279, 2004
|
|
1V8U
 
 | | Crystal structure analysis of the ADP-ribose pyrophosphatase of E82Q mutant with SO4 and Mg | | Descriptor: | ADP-ribose pyrophosphatase, MAGNESIUM ION, SULFATE ION | | Authors: | Yoshiba, S, Ooga, T, Nakagawa, N, Shibata, T, Inoue, Y, Yokoyama, S, Kuramitsu, S, Masui, R, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2004-01-15 | | Release date: | 2004-10-19 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural insights into the Thermus thermophilus ADP-ribose pyrophosphatase mechanism via crystal structures with the bound substrate and metal
J.Biol.Chem., 279, 2004
|
|
1V8V
 
 | | Crystal structure analysis of the ADP-ribose pyrophosphatase of E86Q mutant, complexed with ADP-ribose and Mg | | Descriptor: | ADENOSINE-5-DIPHOSPHORIBOSE, ADP-ribose pyrophosphatase, MAGNESIUM ION | | Authors: | Yoshiba, S, Ooga, T, Nakagawa, N, Shibata, T, Inoue, Y, Yokoyama, S, Kuramitsu, S, Masui, R, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2004-01-15 | | Release date: | 2004-10-19 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | Structural insights into the Thermus thermophilus ADP-ribose pyrophosphatase mechanism via crystal structures with the bound substrate and metal
J.Biol.Chem., 279, 2004
|
|
1V8W
 
 | | Crystal structure analysis of the ADP-ribose pyrophosphatase of E82Q mutant, complexed with SO4 and Zn | | Descriptor: | ADP-ribose pyrophosphatase, SULFATE ION, ZINC ION | | Authors: | Yoshiba, S, Ooga, T, Nakagawa, N, Shibata, T, Inoue, Y, Yokoyama, S, Kuramitsu, S, Masui, R, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2004-01-15 | | Release date: | 2004-10-19 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.66 Å) | | Cite: | Structural insights into the Thermus thermophilus ADP-ribose pyrophosphatase mechanism via crystal structures with the bound substrate and metal
J.Biol.Chem., 279, 2004
|
|
1V8X
 
 | | Crystal Structure of the Dioxygen-bound Heme Oxygenase from Corynebacterium diphtheriae | | Descriptor: | Heme oxygenase, OXYGEN MOLECULE, PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | Unno, M, Matsui, T, Chu, G.C, Couture, M, Yoshida, T, Rousseau, D.L, Olson, J.S, Ikeda-Saito, M. | | Deposit date: | 2004-01-15 | | Release date: | 2004-05-18 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Crystal Structure of the Dioxygen-bound Heme Oxygenase from Corynebacterium diphtheriae: IMPLICATIONS FOR HEME OXYGENASE FUNCTION.
J.Biol.Chem., 279, 2004
|
|
1V8Y
 
 | | Crystal structure analysis of the ADP-ribose pyrophosphatase of E86Q mutant, complexed with ADP-ribose and Zn | | Descriptor: | ADENOSINE-5-DIPHOSPHORIBOSE, ADP-ribose pyrophosphatase, ZINC ION | | Authors: | Yoshiba, S, Ooga, T, Nakagawa, N, Shibata, T, Inoue, Y, Yokoyama, S, Kuramitsu, S, Masui, R, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2004-01-15 | | Release date: | 2004-10-19 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Structural insights into the Thermus thermophilus ADP-ribose pyrophosphatase mechanism via crystal structures with the bound substrate and metal
J.Biol.Chem., 279, 2004
|
|
1V8Z
 
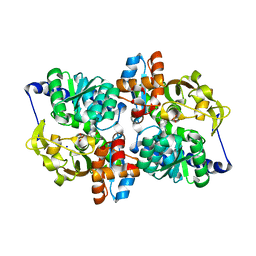 | | X-ray crystal structure of the Tryptophan Synthase b2 Subunit from Hyperthermophile, Pyrococcus furiosus | | Descriptor: | PYRIDOXAL-5'-PHOSPHATE, SODIUM ION, Tryptophan synthase beta chain 1 | | Authors: | Hioki, Y, Ogasahara, K, Lee, S.J, Ma, J, Ishida, M, Yamagata, Y, Matsuura, Y, Ota, M, Kuramitsu, S, Yutani, K, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2004-01-15 | | Release date: | 2005-02-22 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.21 Å) | | Cite: | The crystal structure of the tryptophan synthase beta subunit from the hyperthermophile Pyrococcus furiosus. Investigation of stabilization factors
Eur.J.Biochem., 271, 2004
|
|
1V90
 
 | | Solution structure by NMR means of delta-paluIT1-NH2 | | Descriptor: | Delta-palutoxin IT1 | | Authors: | Ferrat, G, Bosmans, F, Tytgat, J, Pimentel, C, Chagot, B, Nakajima, T, Darbon, H, Corzo, G. | | Deposit date: | 2004-01-19 | | Release date: | 2005-03-29 | | Last modified: | 2024-10-16 | | Method: | SOLUTION NMR | | Cite: | Solution structure of two insect-specific spider toxins and their pharmacological interaction with the insect voltage-gated Na(+) channel
Proteins, 59, 2005
|
|
1V91
 
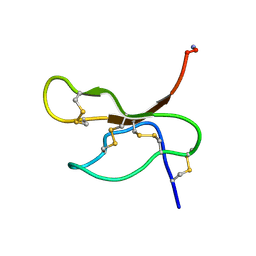 | | Solution structure of insectidal toxin delta-paluIT2-NH2 | | Descriptor: | Delta-palutoxin IT2 | | Authors: | Ferrat, G, Bosmans, F, Tytgat, J, Pimentel, C, Chagot, B, Nakajima, T, Darbon, H, Corzo, G. | | Deposit date: | 2004-01-19 | | Release date: | 2005-03-29 | | Last modified: | 2024-11-06 | | Method: | SOLUTION NMR | | Cite: | Solution structure of two insect-specific spider toxins and their pharmacological interaction with the insect voltage-gated Na(+) channel
Proteins, 59, 2005
|
|
1V92
 
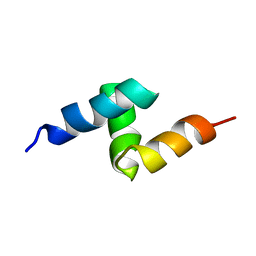 | | Solution structure of the UBA domain from p47, a major cofactor of the AAA ATPase p97 | | Descriptor: | NSFL1 cofactor p47 | | Authors: | Yuan, X, Simpson, P, Mckeown, C, Kondo, H, Uchiyama, K, Wallis, R, Dreveny, I, Keetch, C, Zhang, X, Robinson, C, Freemont, P, Matthews, S. | | Deposit date: | 2004-01-19 | | Release date: | 2004-04-06 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | Structure, dynamics and interactions of p47, a major adaptor of the AAA ATPase, p97
Embo J., 23, 2004
|
|
1V93
 
 | | 5,10-Methylenetetrahydrofolate Reductase from Thermus thermophilus HB8 | | Descriptor: | 1,4-DIETHYLENE DIOXIDE, 5,10-Methylenetetrahydrofolate reductase, FLAVIN-ADENINE DINUCLEOTIDE | | Authors: | Nakajima, Y, Miyahara, I, Hirotsu, K, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2004-01-20 | | Release date: | 2004-02-03 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal Structure of 5,10-Methylenetetrahydrofolate reductase from Thermus thermophilus HB8
To be Published
|
|
1V94
 
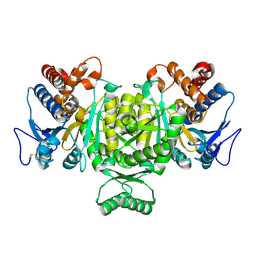 | | Crystal structure of isocitrate dehydrogenase from Aeropyrum pernix | | Descriptor: | isocitrate dehydrogenase | | Authors: | Jeong, J.-J, Sonoda, T, Fushinobu, S, Shoun, H, Wakagi, T. | | Deposit date: | 2004-01-20 | | Release date: | 2005-01-25 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.28 Å) | | Cite: | Crystal structure of isocitrate dehydrogenase from Aeropyrum pernix
Proteins, 55, 2004
|
|
1V95
 
 | | Solution Structure of Anticodon Binding Domain from Nuclear Receptor Coactivator 5 (Human KIAA1637 Protein) | | Descriptor: | Nuclear receptor coactivator 5 | | Authors: | Zhao, C, Kigawa, T, Tochio, N, Koshiba, S, Inoue, M, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2004-01-20 | | Release date: | 2004-07-20 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | Solution Structure of Anticodon Binding Domain from Nuclear Receptor Coactivator 5 (Human KIAA1637 Protein)
To be Published
|
|
1V96
 
 | |
1V97
 
 | | Crystal Structure of Bovine Milk Xanthine Dehydrogenase FYX-051 bound form | | Descriptor: | 4-(5-PYRIDIN-4-YL-1H-1,2,4-TRIAZOL-3-YL)PYRIDINE-2-CARBONITRILE, ACETIC ACID, CALCIUM ION, ... | | Authors: | Okamoto, K, Matsumoto, K, Hille, R, Eger, B.T, Pai, E.F, Nishino, T. | | Deposit date: | 2004-01-21 | | Release date: | 2004-06-22 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | The crystal structure of xanthine oxidoreductase during catalysis: Implications for reaction mechanism and enzyme inhibition.
Proc.Natl.Acad.Sci.USA, 101, 2004
|
|
1V98
 
 | |
1V9A
 
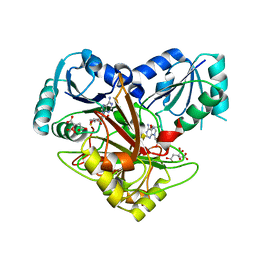 | | Crystal structure of Uroporphyrin-III C-methyl transferase from Thermus thermophilus complexed with S-adenyl homocysteine | | Descriptor: | CITRATE ANION, S-ADENOSYL-L-HOMOCYSTEINE, Uroporphyrin-III C-methyltransferase | | Authors: | Rehse, P.H, Kitao, T, Tahirov, T.H, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2004-01-23 | | Release date: | 2005-02-01 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure of a closed-form uroporphyrinogen-III C-methyltransferase from Thermus thermophilus.
Acta Crystallogr.,Sect.D, 61, 2005
|
|
1V9C
 
 | |
1V9D
 
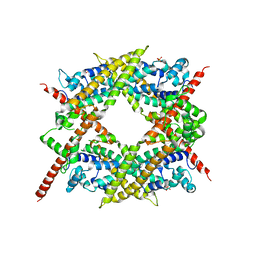 | | Crystal structure of the core FH2 domain of mouse mDia1 | | Descriptor: | Diaphanous protein homolog 1, SULFATE ION | | Authors: | Shimada, A, Nyitrai, M, Vetter, I.R, Kuhlmann, D, Bugyi, B, Narumiya, S, Geeves, M.A, Wittinghofer, A. | | Deposit date: | 2004-01-24 | | Release date: | 2004-03-09 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | The core FH2 domain of diaphanous-related formins is an elongated actin binding protein that inhibits polymerization.
Mol.Cell, 13, 2004
|
|
1V9E
 
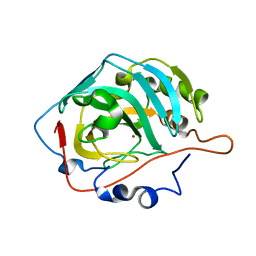 | | Crystal Structure Analysis of Bovine Carbonic Anhydrase II | | Descriptor: | Carbonic anhydrase II, ZINC ION | | Authors: | Saito, R, Sato, T, Ikai, A, Tanaka, N. | | Deposit date: | 2004-01-26 | | Release date: | 2004-02-10 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structure of bovine carbonic anhydrase II at 1.95 A resolution.
Acta Crystallogr.,Sect.D, 60, 2004
|
|
1V9F
 
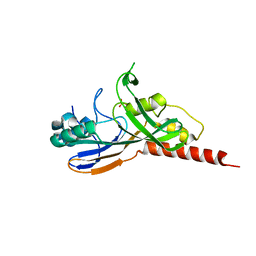 | | Crystal structure of catalytic domain of pseudouridine synthase RluD from Escherichia coli | | Descriptor: | PHOSPHATE ION, Ribosomal large subunit pseudouridine synthase D | | Authors: | Mizutani, K, Machida, Y, Unzai, S, Park, S.-Y, Tame, J.R.H. | | Deposit date: | 2004-01-26 | | Release date: | 2004-05-18 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structures of the catalytic domains of pseudouridine synthases RluC and RluD from Escherichia coli
Biochemistry, 43, 2004
|
|
1V9G
 
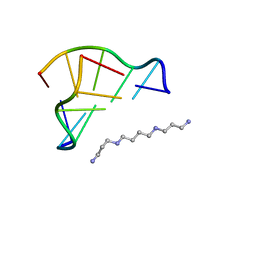 | | Neutron Crystallographic analysis of the Z-DNA hexamer CGCGCG | | Descriptor: | 5'-D(*CP*GP*CP*GP*CP*G)-3', N,N'-BIS(3-AMMONIOPROPYL)BUTANE-1,4-DIAMINIUM | | Authors: | Chatake, T, Tanaka, I, Niimura, N. | | Deposit date: | 2004-01-26 | | Release date: | 2005-01-26 | | Last modified: | 2023-10-25 | | Method: | NEUTRON DIFFRACTION (1.8 Å) | | Cite: | The hydration structure of a Z-DNA hexameric duplex determined by a neutron diffraction technique.
Acta Crystallogr.,Sect.D, 61, 2005
|
|
