1KXD
 
 | |
8VJP
 
 | | Histidine-covalent stapled alpha-helical peptide (155H1) targeting hMcl-1 | | Descriptor: | (4Z)-oct-4-en-1-ol, (S~1~R)-3-carbamoyl-4-methoxybenzene-1-sulfinic acid, Histidine-covalent stapled alpha-helical peptide, ... | | Authors: | Muzzarelli, K.M, Assar, Z, Alboreggia, G, Pellecchia, M. | | Deposit date: | 2024-01-07 | | Release date: | 2024-05-15 | | Last modified: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (1.13 Å) | | Cite: | Histidine-Covalent Stapled Alpha-Helical Peptides Targeting hMcl-1.
J.Med.Chem., 67, 2024
|
|
1UX7
 
 | | Carbohydrate-Binding Module CBM36 in complex with calcium and xylotriose | | Descriptor: | CALCIUM ION, ENDO-1,4-BETA-XYLANASE D, SULFATE ION, ... | | Authors: | Davies, G.J, Boraston, A.B, Jamal, S. | | Deposit date: | 2004-02-19 | | Release date: | 2004-10-27 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Ab Initio Structure Determination and Functional Characterization of Cbm36: A New Family of Calcium-Dependent Carbohydrate Binding Modules
Structure, 12, 2004
|
|
1KXE
 
 | |
8V38
 
 | |
1Z6Z
 
 | | Crystal Structure of Human Sepiapterin Reductase in complex with NADP+ | | Descriptor: | CHLORIDE ION, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, SULFATE ION, ... | | Authors: | Ugochukwu, E, Kavanagh, K, Ng, S, Arrowsmith, C, Edwards, A, Sundstrom, M, von Delft, F, Oppermann, U, Structural Genomics Consortium (SGC) | | Deposit date: | 2005-03-23 | | Release date: | 2005-04-05 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal Structure of Human Sepiapterin Reductase
To be Published
|
|
2WZH
 
 | | BtGH84 D242N in complex with MeUMB-derived oxazoline | | Descriptor: | 2-METHYL-4,5-DIHYDRO-(1,2-DIDEOXY-ALPHA-D-GLUCOPYRANOSO)[2,1-D]-1,3-OXAZOLE, CALCIUM ION, GLYCEROL, ... | | Authors: | He, Y, Davies, G.J. | | Deposit date: | 2009-11-30 | | Release date: | 2010-01-26 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Visualizing the Reaction Coordinate of an O-Glcnac Hydrolase
J.Am.Chem.Soc., 132, 2010
|
|
8VE9
 
 | | IsPETase - ACCCETN mutant - CombiPETase | | Descriptor: | CHLORIDE ION, GLYCEROL, Poly(ethylene terephthalate) hydrolase | | Authors: | Joho, Y, Royan, S, Newton, S, Caputo, A.T, Ardevol Grau, A, Jackson, C. | | Deposit date: | 2023-12-18 | | Release date: | 2024-06-19 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Enhancing PET Degrading Enzymes: A Combinatory Approach.
Chembiochem, 25, 2024
|
|
1SWU
 
 | | STREPTAVIDIN MUTANT Y43F | | Descriptor: | (4R)-2-METHYLPENTANE-2,4-DIOL, (4S)-2-METHYL-2,4-PENTANEDIOL, STREPTAVIDIN | | Authors: | Freitag, S, Le Trong, I, Klumb, L.A, Stayton, P.S, Stenkamp, R.E. | | Deposit date: | 1998-10-12 | | Release date: | 1999-11-10 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.14 Å) | | Cite: | Atomic resolution structure of biotin-free Tyr43Phe streptavidin: what is in the binding site?
Acta Crystallogr.,Sect.D, 55, 1999
|
|
1YWU
 
 | | Solution NMR structure of Pseudomonas Aeruginosa protein PA4608. Northeast Structural Genomics target PaT7 | | Descriptor: | hypothetical protein PA4608 | | Authors: | Ramelot, T.A, Yee, A.A, Cort, J.R, Semesi, A, Arrowsmith, C.H, Kennedy, M.A, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2005-02-18 | | Release date: | 2005-03-29 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | NMR structure and binding studies confirm that PA4608 from Pseudomonas aeruginosa is a PilZ domain and a c-di-GMP binding protein.
Proteins, 66, 2007
|
|
1Z82
 
 | |
2WXU
 
 | | Clostridium perfringens alpha-toxin strain NCTC8237 mutant T74I | | Descriptor: | CADMIUM ION, CALCIUM ION, PHOSPHOLIPASE C, ... | | Authors: | Vachieri, S.G, Naylor, C.E, Basak, A.K. | | Deposit date: | 2009-11-10 | | Release date: | 2009-11-17 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Comparison of a Nontoxic Variant of Clostridium Perfringens [Alpha]-Toxin with the Toxic Wild-Type Strain
Acta Crystallogr.,Sect.D, 66, 2010
|
|
8V0T
 
 | |
8VKJ
 
 | | Cryo-EM structure of human HGSNAT bound with Acetyl-CoA | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ACETYL COENZYME *A, ... | | Authors: | Li, F, Zhao, B. | | Deposit date: | 2024-01-09 | | Release date: | 2024-06-26 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (2.92 Å) | | Cite: | Structural and mechanistic insights into a lysosomal membrane enzyme HGSNAT involved in Sanfilippo syndrome.
Nat Commun, 15, 2024
|
|
8VQY
 
 | | Human GABAA receptor alpha1-beta2-gamma2 subtype in complex with GABA plus methaqualone | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-methyl-3-(2-methylphenyl)quinazolin-4(3H)-one, GAMMA-AMINO-BUTANOIC ACID, ... | | Authors: | Chojnacka, W, Teng, J, Kim, J.J, Jensen, A.A, Hibbs, R.E. | | Deposit date: | 2024-01-19 | | Release date: | 2024-06-26 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (2.82 Å) | | Cite: | Structural insights into GABA A receptor potentiation by Quaalude.
Nat Commun, 15, 2024
|
|
7BIK
 
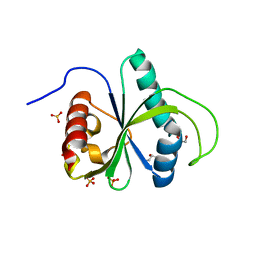 | | Crystal structure of YTHDF2 in complex with m6Am | | Descriptor: | (2~{R},3~{S},4~{R},5~{R})-2-(hydroxymethyl)-4-methoxy-5-[6-(methylamino)purin-9-yl]oxolan-3-ol, GLYCEROL, SULFATE ION, ... | | Authors: | Wang, X, Caflisch, A. | | Deposit date: | 2021-01-12 | | Release date: | 2021-11-24 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure of YTHDF2 in complex with m6Am
To Be Published
|
|
8W0S
 
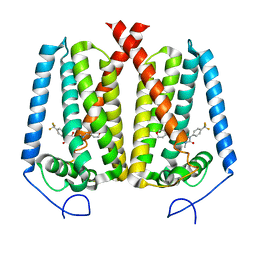 | | Human EBP complexed with compound 3a | | Descriptor: | 1-methyl-8-[(oxan-4-yl)methyl]-3-[4-(trifluoromethyl)phenyl]-1,3,8-triazaspiro[4.5]decane-2,4-dione, 3-beta-hydroxysteroid-Delta(8),Delta(7)-isomerase | | Authors: | Sun, D, Masureel, M. | | Deposit date: | 2024-02-14 | | Release date: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Discovery and Optimization of Selective Brain-Penetrant EBP Inhibitors that Enhance Oligodendrocyte Formation.
J.Med.Chem., 67, 2024
|
|
8VLV
 
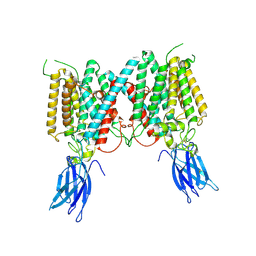 | | Cryo-EM structure of human HGSNAT in inactive state | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Heparan-alpha-glucosaminide N-acetyltransferase | | Authors: | Li, F. | | Deposit date: | 2024-01-12 | | Release date: | 2024-06-26 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.49 Å) | | Cite: | Structural and mechanistic insights into a lysosomal membrane enzyme HGSNAT involved in Sanfilippo syndrome.
Nat Commun, 15, 2024
|
|
1Z8U
 
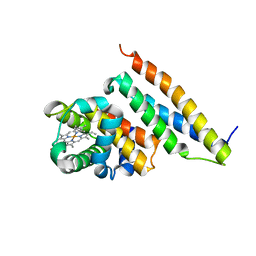 | | Crystal structure of oxidized alpha hemoglobin bound to AHSP | | Descriptor: | Alpha-hemoglobin stabilizing protein, Hemoglobin alpha chain, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Feng, L, Zhou, S, Gu, L, Gell, D.A, Mackay, J.P, Weiss, M.J, Gow, A.J, Shi, Y. | | Deposit date: | 2005-03-31 | | Release date: | 2005-06-14 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structure of oxidized alpha-haemoglobin bound to AHSP reveals a protective mechanism for haem.
Nature, 435, 2005
|
|
8VRN
 
 | | Human GABAA receptor alpha1-beta2-gamma2 subtype in complex with GABA plus PPTQ | | Descriptor: | (2S)-3-(hexadecanoyloxy)-2-[(9Z)-octadec-9-enoyloxy]propyl 2-(trimethylammonio)ethyl phosphate, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 3-(4-methylphenyl)-2-phenylquinazolin-4(3H)-one, ... | | Authors: | Chojnacka, W, Teng, J, Kim, J.J, Jensen, A.A, Hibbs, R.E. | | Deposit date: | 2024-01-22 | | Release date: | 2024-06-26 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (2.57 Å) | | Cite: | Structural insights into GABA A receptor potentiation by Quaalude.
Nat Commun, 15, 2024
|
|
1Z97
 
 | | Human Carbonic Anhydrase III: Structural and Kinetic Study of Catalysis and Proton Transfer. | | Descriptor: | Carbonic anhydrase III, ZINC ION | | Authors: | Duda, D.M, Tu, C, Fisher, S.Z, An, H, Yoshioka, C, Govindasamy, L, Laipis, P.J, Agbandje-McKenna, M, Silverman, D.N, McKenna, R. | | Deposit date: | 2005-03-31 | | Release date: | 2005-08-09 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Human Carbonic Anhydrase III: Structural and Kinetic Study of Catalysis and Proton Transfer
Biochemistry, 44, 2005
|
|
8V0P
 
 | | SARS-CoV-2 Omicron-XBB.1.16 3-RBD down Spike Protein Trimer 2 (S-GSAS-Omicron-XBB.1.16) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein | | Authors: | Zhang, Q.E, Acharya, P. | | Deposit date: | 2023-11-17 | | Release date: | 2024-06-12 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | SARS-CoV-2 Omicron XBB lineage spike structures, conformations, antigenicity, and receptor recognition.
Mol.Cell, 84, 2024
|
|
1KXF
 
 | |
8V0O
 
 | | SARS-CoV-2 Omicron-XBB.1.16 3-RBD down Spike Protein Trimer 1 (S-GSAS-Omicron-XBB.1.16) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein | | Authors: | Zhang, Q.E, Acharya, P. | | Deposit date: | 2023-11-17 | | Release date: | 2024-06-12 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | SARS-CoV-2 Omicron XBB lineage spike structures, conformations, antigenicity, and receptor recognition.
Mol.Cell, 84, 2024
|
|
7BGI
 
 | | Photosystem I of a temperature sensitive mutant Chlamydomonas reinhardtii | | Descriptor: | (1~{S})-3,5,5-trimethyl-4-[(1~{E},3~{E},5~{E},7~{E},9~{E},11~{E},13~{E},15~{E},17~{E})-3,7,12,16-tetramethyl-18-[(4~{S})-2,6,6-trimethyl-4-oxidanyl-cyclohexen-1-yl]octadeca-1,3,5,7,9,11,13,15,17-nonaenyl]cyclohex-3-en-1-ol, (2R)-2-hydroxy-3-(phosphonooxy)propyl (9E)-octadec-9-enoate, (2S)-3-{[(R)-(2-aminoethoxy)(hydroxy)phosphoryl]oxy}-2-hydroxypropyl hexadecanoate, ... | | Authors: | Caspy, I, Nelson, N. | | Deposit date: | 2021-01-07 | | Release date: | 2021-12-08 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (2.54 Å) | | Cite: | Dimeric and high-resolution structures of Chlamydomonas Photosystem I from a temperature-sensitive Photosystem II mutant
Commun Biol, 4, 2021
|
|
