3AZS
 
 | | Diverse Substrates Recognition Mechanism Revealed by Thermotoga maritima Cel5A Structures in Complex with Mannotriose | | Descriptor: | Endoglucanase, beta-D-mannopyranose-(1-4)-beta-D-mannopyranose-(1-4)-alpha-D-mannopyranose | | Authors: | Wu, T.H, Huang, C.H, Ko, T.P, Lai, H.L, Ma, Y, Chen, C.C, Cheng, Y.S, Liu, J.R, Guo, R.T. | | Deposit date: | 2011-05-30 | | Release date: | 2011-08-10 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.69 Å) | | Cite: | Diverse substrate recognition mechanism revealed by Thermotoga maritima Cel5A structures in complex with cellotetraose, cellobiose and mannotriose
Biochim.Biophys.Acta, 1814, 2011
|
|
3AZY
 
 | | Crystal structure of the laminarinase catalytic domain from Thermotoga maritima MSB8 | | Descriptor: | CALCIUM ION, Laminarinase, SULFATE ION | | Authors: | Jeng, W.Y, Wang, N.C, Wang, A.H.J. | | Deposit date: | 2011-06-03 | | Release date: | 2011-11-23 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Crystal structures of the laminarinase catalytic domain from Thermotoga maritima MSB8 in complex with inhibitors: essential residues for beta-1,3 and beta-1,4 glucan selection.
J.Biol.Chem., 286, 2011
|
|
3AZT
 
 | | Diverse Substrates Recognition Mechanism Revealed by Thermotoga maritima Cel5A Structures in Complex with Cellotetraose | | Descriptor: | Endoglucanase, beta-D-glucopyranose-(1-4)-beta-D-glucopyranose-(1-4)-beta-D-glucopyranose-(1-4)-alpha-D-glucopyranose | | Authors: | Wu, T.H, Huang, C.H, Ko, T.P, Lai, H.L, Ma, Y, Chen, C.C, Cheng, Y.S, Liu, J.R, Guo, R.T. | | Deposit date: | 2011-05-30 | | Release date: | 2011-08-10 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Diverse substrate recognition mechanism revealed by Thermotoga maritima Cel5A structures in complex with cellotetraose, cellobiose and mannotriose
Biochim.Biophys.Acta, 1814, 2011
|
|
3B00
 
 | | Crystal structure of the laminarinase catalytic domain from Thermotoga maritima MSB8 in complex with cetyltrimethylammonium bromide | | Descriptor: | CALCIUM ION, CETYL-TRIMETHYL-AMMONIUM, Laminarinase | | Authors: | Jeng, W.Y, Wang, N.C, Wang, A.H.J. | | Deposit date: | 2011-06-03 | | Release date: | 2011-11-23 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.74 Å) | | Cite: | Crystal structures of the laminarinase catalytic domain from Thermotoga maritima MSB8 in complex with inhibitors: essential residues for beta-1,3 and beta-1,4 glucan selection.
J.Biol.Chem., 286, 2011
|
|
2GAI
 
 | |
2K3F
 
 | |
2GAJ
 
 | |
2ISS
 
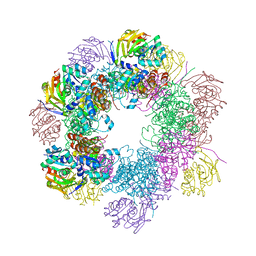 | | Structure of the PLP synthase Holoenzyme from Thermotoga maritima | | Descriptor: | Glutamine amidotransferase subunit pdxT, PHOSPHATE ION, Pyridoxal biosynthesis lyase pdxS, ... | | Authors: | Zein, F, Zhang, Y, Kang, Y.N, Burns, K, Begley, T.P, Ealick, S.E. | | Deposit date: | 2006-10-18 | | Release date: | 2007-01-02 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structural Insights into the Mechanism of the PLP Synthase Holoenzyme from Thermotoga maritima
Biochemistry, 45, 2006
|
|
1NF2
 
 | |
2J71
 
 | |
3AMP
 
 | | E134C-Cellotetraose complex of cellulase 12A from thermotoga maritima | | Descriptor: | Endo-1,4-beta-glucanase, beta-D-glucopyranose-(1-4)-beta-D-glucopyranose, beta-D-glucopyranose-(1-4)-beta-D-glucopyranose-(1-4)-alpha-D-glucopyranose | | Authors: | Cheng, Y.-S, Ko, T.-P, Liu, J.-R, Guo, R.-T. | | Deposit date: | 2010-08-20 | | Release date: | 2011-03-16 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | Crystal structure and substrate-binding mode of cellulase 12A from Thermotoga maritima
Proteins, 79, 2011
|
|
2J72
 
 | |
1MKM
 
 | | CRYSTAL STRUCTURE OF THE THERMOTOGA MARITIMA ICLR | | Descriptor: | FORMIC ACID, IclR transcriptional regulator, ZINC ION | | Authors: | Kim, Y, Zhang, R.G, Joachimiak, A, Skarina, T, Edwards, A, Savchenko, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2002-08-29 | | Release date: | 2002-09-11 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of Thermotoga maritima 0065, a member of the IclR transcriptional factor family.
J.Biol.Chem., 277, 2002
|
|
3AMH
 
 | | crystal structure of cellulase 12A from Thermotoga maritima | | Descriptor: | Endo-1,4-beta-glucanase | | Authors: | Cheng, Y.-S, Ko, T.-P, Liu, J.-R, Guo, R.-T. | | Deposit date: | 2010-08-20 | | Release date: | 2011-03-16 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.09 Å) | | Cite: | Crystal structure and substrate-binding mode of cellulase 12A from Thermotoga maritima
Proteins, 79, 2011
|
|
3AMQ
 
 | | E134C-Cellobiose co-crystal of cellulase 12A from thermotoga maritima | | Descriptor: | Endo-1,4-beta-glucanase, beta-D-glucopyranose, beta-D-glucopyranose-(1-4)-alpha-D-glucopyranose | | Authors: | Cheng, Y.-S, Ko, T.-P, Liu, J.-R, Guo, R.-T. | | Deposit date: | 2010-08-20 | | Release date: | 2011-03-16 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure and substrate-binding mode of cellulase 12A from Thermotoga maritima
Proteins, 79, 2011
|
|
3AMM
 
 | | Cellotetraose complex of cellulase 12A from thermotoga maritima | | Descriptor: | Endo-1,4-beta-glucanase, beta-D-glucopyranose-(1-4)-beta-D-glucopyranose-(1-4)-beta-D-glucopyranose-(1-4)-beta-D-glucopyranose | | Authors: | Cheng, Y.-S, Ko, T.-P, Liu, J.-R, Guo, R.-T. | | Deposit date: | 2010-08-20 | | Release date: | 2011-03-16 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | Crystal structure and substrate-binding mode of cellulase 12A from Thermotoga maritima
Proteins, 79, 2011
|
|
3AMN
 
 | | E134C-Cellobiose complex of cellulase 12A from thermotoga maritima | | Descriptor: | Endo-1,4-beta-glucanase, beta-D-glucopyranose-(1-4)-alpha-D-glucopyranose, beta-D-glucopyranose-(1-4)-beta-D-glucopyranose | | Authors: | Cheng, Y.-S, Ko, T.-P, Liu, J.-R, Guo, R.-T. | | Deposit date: | 2010-08-20 | | Release date: | 2011-03-16 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.47 Å) | | Cite: | Crystal structure and substrate-binding mode of cellulase 12A from Thermotoga maritima
Proteins, 79, 2011
|
|
1NZ0
 
 | | RNASE P PROTEIN FROM THERMOTOGA MARITIMA | | Descriptor: | Ribonuclease P protein component, SULFATE ION | | Authors: | Kazantsev, A.V, Krivenko, A.A, Harrington, D.J, Carter, R.J, Holbrook, S.R, Adams, P.D, Pace, N.R, Berkeley Structural Genomics Center (BSGC) | | Deposit date: | 2003-02-14 | | Release date: | 2003-06-24 | | Last modified: | 2021-10-27 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | High-resolution structure of RNase P protein from Thermotoga maritima.
Proc.Natl.Acad.Sci.USA, 100, 2003
|
|
2H3H
 
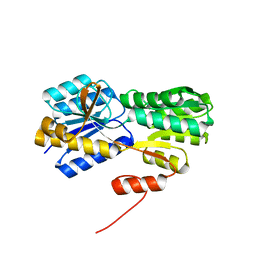 | |
1M4Y
 
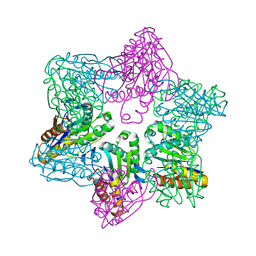 | | Crystal structure of HslV from Thermotoga maritima | | Descriptor: | ATP-dependent protease hslV, SODIUM ION | | Authors: | Song, H.K, Ramachandran, R, Bochtler, M.B, Hartmann, C, Azim, M.K, Huber, R. | | Deposit date: | 2002-07-05 | | Release date: | 2003-05-06 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Isolation and characterization of the prokaryotic proteasome homolog HslVU (ClpQY) from Thermotoga maritima and the crystal structure of HslV.
BIOPHYS.CHEM., 100, 2003
|
|
1MGP
 
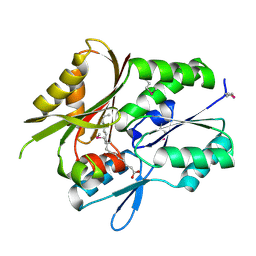 | | Hypothetical protein TM841 from Thermotoga maritima reveals fatty acid binding function | | Descriptor: | Hypothetical protein TM841, PALMITIC ACID | | Authors: | Schulze-Gahmen, U, Pelaschier, J, Yokota, H, Kim, R, Kim, S.-H, Berkeley Structural Genomics Center (BSGC) | | Deposit date: | 2002-08-15 | | Release date: | 2002-09-18 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of a hypothetical protein, TM841 of Thermotoga maritima,
reveals its function as fatty acid binding protein
Proteins, 50, 2003
|
|
3AFH
 
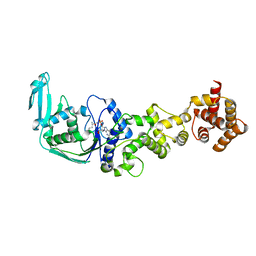 | |
1OBB
 
 | | alpha-glucosidase A, AglA, from Thermotoga maritima in complex with maltose and NAD+ | | Descriptor: | ALPHA-GLUCOSIDASE, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose | | Authors: | Lodge, J.A, Maier, T, Liebl, W, Hoffmann, V, Strater, N. | | Deposit date: | 2003-01-29 | | Release date: | 2003-05-21 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal Structure of Thermotoga Maritima Alpha-Glucosidase Agla Defines a New Clan of Nad+-Dependent Glycosidases
J.Biol.Chem., 278, 2003
|
|
1SAZ
 
 | | Membership in the ASKHA Superfamily: Enzymological Properties and Crystal Structure of Butyrate Kinase 2 from Thermotoga maritima | | Descriptor: | FORMIC ACID, PHOSPHOMETHYLPHOSPHONIC ACID ADENYLATE ESTER, Probable butyrate kinase 2, ... | | Authors: | Diao, J, Cooper, D.R, Sanders, D.A, Hasson, M.S. | | Deposit date: | 2004-02-09 | | Release date: | 2005-03-29 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of butyrate kinase 2 from Thermotoga maritima, a member of the ASKHA superfamily of phosphotransferases.
J.Bacteriol., 191, 2009
|
|
1V4E
 
 | | Crystal Structure of Octaprenyl Pyrophosphate Synthase from Hyperthermophilic Thermotoga maritima | | Descriptor: | SULFATE ION, octoprenyl-diphosphate synthase | | Authors: | Guo, R.T, Kuo, C.J, Chou, C.C, Ko, T.P, Shr, H.L, Liang, P.H, Wang, A.H.-J. | | Deposit date: | 2003-11-13 | | Release date: | 2004-03-02 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.28 Å) | | Cite: | Crystal Structure of Octaprenyl Pyrophosphate Synthase from Hyperthermophilic Thermotoga maritima and Mechanism of Product Chain Length Determination
J.Biol.Chem., 279, 2004
|
|
