6D8Q
 
 | |
6D8R
 
 | |
6D8Y
 
 | |
6DRF
 
 | | Structure of human Retinal Degeneration 3(RD3) Protein | | Descriptor: | Protein RD3 | | Authors: | Yu, Q, Lim, S, Peshenko, I, Cudia, D, Dizhoor, A.M, Ames, J.B. | | Deposit date: | 2018-06-11 | | Release date: | 2019-02-06 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Retinal degeneration 3 (RD3) protein, a retinal guanylyl cyclase regulator, forms a monomeric and elongated four-helix bundle.
J. Biol. Chem., 294, 2019
|
|
6D93
 
 | |
6D8U
 
 | |
6D9P
 
 | |
6D8S
 
 | |
6D3T
 
 | |
6DYS
 
 | | C-terminal condensation domain of Ebony in complex with beta-alanyl-dopamine | | Descriptor: | CALCIUM ION, Ebony, N-[2-(3,4-dihydroxyphenyl)ethyl]-beta-alaninamide | | Authors: | Izore, T, Tailhades, J, Hansen, M.H, Kaczmarski, J.A, Jackson, C.J, Cryle, M.J. | | Deposit date: | 2018-07-02 | | Release date: | 2019-01-30 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Drosophila melanogasternonribosomal peptide synthetase Ebony encodes an atypical condensation domain.
Proc. Natl. Acad. Sci. U.S.A., 116, 2019
|
|
6DYN
 
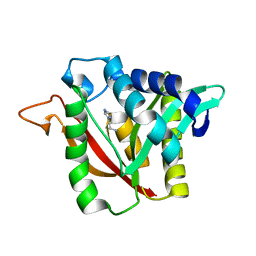 | | C-terminal condensation domain of Ebony in complex with Histamine | | Descriptor: | CALCIUM ION, Ebony, HISTAMINE | | Authors: | Izore, T, Tailhades, J, Hansen, M.H, Kaczmarski, J.A, Jackson, C.J, Cryle, M.J. | | Deposit date: | 2018-07-02 | | Release date: | 2019-01-30 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.102 Å) | | Cite: | Drosophila melanogasternonribosomal peptide synthetase Ebony encodes an atypical condensation domain.
Proc. Natl. Acad. Sci. U.S.A., 116, 2019
|
|
6DZ9
 
 | |
6DZB
 
 | |
6DYO
 
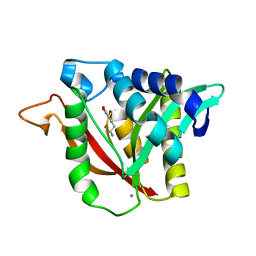 | | C-terminal condensation domain of Ebony in complex with L-Dopamine | | Descriptor: | CALCIUM ION, Ebony, L-DOPAMINE | | Authors: | Izore, T, Tailhades, J, Hansen, M.H, Kaczmarski, J.A, Jackson, C.J, Cryle, M.J. | | Deposit date: | 2018-07-02 | | Release date: | 2019-01-30 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.84 Å) | | Cite: | Drosophila melanogasternonribosomal peptide synthetase Ebony encodes an atypical condensation domain.
Proc. Natl. Acad. Sci. U.S.A., 116, 2019
|
|
6DYM
 
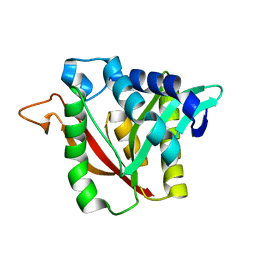 | | C-terminal condensation domain of Ebony | | Descriptor: | CALCIUM ION, Ebony | | Authors: | Izore, T, Tailhades, J, Hansen, M.H, Kaczmarski, J.A, Jackson, C.J, Cryle, M.J. | | Deposit date: | 2018-07-02 | | Release date: | 2019-01-30 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.02 Å) | | Cite: | Drosophila melanogasternonribosomal peptide synthetase Ebony encodes an atypical condensation domain.
Proc. Natl. Acad. Sci. U.S.A., 116, 2019
|
|
6DYR
 
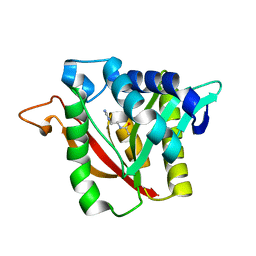 | | C-terminal condensation domain of Ebony in complex with Carcinine | | Descriptor: | Ebony, N-[2-(1H-imidazol-5-yl)ethyl]-beta-alaninamide | | Authors: | Izore, T, Tailhades, J, Hansen, M.H, Kaczmarski, J.A, Jackson, C.J, Cryle, M.J. | | Deposit date: | 2018-07-02 | | Release date: | 2019-01-30 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Drosophila melanogasternonribosomal peptide synthetase Ebony encodes an atypical condensation domain.
Proc. Natl. Acad. Sci. U.S.A., 116, 2019
|
|
6SFW
 
 | |
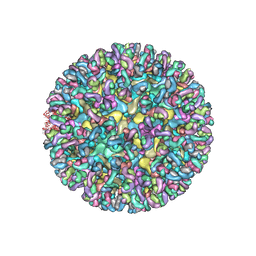
6MW9
 
 | | CryoEM structure of chimeric Eastern Equine Encephalitis Virus with Fab of EEEV-3 antibody | | Descriptor: | E1, E2, EEEV-3 antibody heavy chain, ... | | Authors: | Hasan, S.S, Sun, C, Kim, A.S, Watanabe, Y, Chen, C.L, Klose, T, Buda, G, Crispin, M, Diamond, M.S, Klimstra, W.B, Rossmann, M.G. | | Deposit date: | 2018-10-29 | | Release date: | 2018-12-19 | | Last modified: | 2019-12-18 | | Method: | ELECTRON MICROSCOPY (7.3 Å) | | Cite: | Cryo-EM Structures of Eastern Equine Encephalitis Virus Reveal Mechanisms of Virus Disassembly and Antibody Neutralization.
Cell Rep, 25, 2018
|
|
6SFX
 
 | |
8C8R
 
 | |
8C8O
 
 | |
6PCI
 
 | | EBOV GPdMuc (Makona) in complex with rEBOV-520 and rEBOV-548 Fabs | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Virion spike glycoprotein, Virion spike glycoprotein,Virion spike glycoprotein,Ebola Virus (Makona) GP2, ... | | Authors: | Ward, A.B, Murin, C.D, Alkutkar, T. | | Deposit date: | 2019-06-17 | | Release date: | 2020-03-11 | | Last modified: | 2020-07-29 | | Method: | ELECTRON MICROSCOPY (4.12 Å) | | Cite: | Analysis of a Therapeutic Antibody Cocktail Reveals Determinants for Cooperative and Broad Ebolavirus Neutralization.
Immunity, 52, 2020
|
|
6MUI
 
 | | CryoEM structure of chimeric Eastern Equine Encephalitis Virus with Fab of EEEV-42 antibody | | Descriptor: | E1, E2, EEEV-42 antibody heavy chain, ... | | Authors: | Hasan, S.S, Sun, C, Kim, A.S, Watanabe, Y, Chen, C.L, Klose, T, Buda, G, Crispin, M, Diamond, M.S, Klimstra, W.B, Rossmann, M.G. | | Deposit date: | 2018-10-23 | | Release date: | 2018-12-19 | | Last modified: | 2019-12-18 | | Method: | ELECTRON MICROSCOPY (7.7 Å) | | Cite: | Cryo-EM Structures of Eastern Equine Encephalitis Virus Reveal Mechanisms of Virus Disassembly and Antibody Neutralization.
Cell Rep, 25, 2018
|
|
6TJ3
 
 | |
6MW3
 
 | | EM structure of Bacillus subtilis ribonucleotide reductase inhibited filament composed of NrdE alpha subunit and NrdF beta subunit with dATP | | Descriptor: | 2'-DEOXYADENOSINE 5'-TRIPHOSPHATE, Ribonucleoside-diphosphate reductase, Ribonucleoside-diphosphate reductase NrdF beta subunit | | Authors: | Thomas, W.C, Bacik, J.P, Kaelber, J.T, Ando, N. | | Deposit date: | 2018-10-29 | | Release date: | 2019-06-19 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (4.65 Å) | | Cite: | Convergent allostery in ribonucleotide reductase.
Nat Commun, 10, 2019
|
|
