1BAE
 
 | |
1BAF
 
 | | 2.9 ANGSTROMS RESOLUTION STRUCTURE OF AN ANTI-DINITROPHENYL-SPIN-LABEL MONOCLONAL ANTIBODY FAB FRAGMENT WITH BOUND HAPTEN | | Descriptor: | IGG1-KAPPA AN02 FAB (HEAVY CHAIN), IGG1-KAPPA AN02 FAB (LIGHT CHAIN), N-(2-AMINO-ETHYL)-4,6-DINITRO-N'-(2,2,6,6-TETRAMETHYL-1-OXY-PIPERIDIN-4-YL)-BENZENE-1,3-DIAMINE | | Authors: | Leahy, D.J, Brunger, A.T, Fox, R.O, Hynes, T.R. | | Deposit date: | 1992-01-16 | | Release date: | 1994-01-31 | | Last modified: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | 2.9 A resolution structure of an anti-dinitrophenyl-spin-label monoclonal antibody Fab fragment with bound hapten.
J.Mol.Biol., 221, 1991
|
|
1BAG
 
 | | ALPHA-AMYLASE FROM BACILLUS SUBTILIS COMPLEXED WITH MALTOPENTAOSE | | Descriptor: | ALPHA-1,4-GLUCAN-4-GLUCANOHYDROLASE, CALCIUM ION, alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-beta-D-glucopyranose | | Authors: | Fujimoto, Z, Mizuno, H, Takase, K, Doui, N. | | Deposit date: | 1998-01-30 | | Release date: | 1998-10-21 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of a catalytic-site mutant alpha-amylase from Bacillus subtilis complexed with maltopentaose.
J.Mol.Biol., 277, 1998
|
|
1BAH
 
 | | A TWO DISULFIDE DERIVATIVE OF CHARYBDOTOXIN WITH DISULFIDE 13-33 REPLACED BY TWO ALPHA-AMINOBUTYRIC ACIDS, NMR, 30 STRUCTURES | | Descriptor: | CHARYBDOTOXIN | | Authors: | Song, J, Gilquin, B, Jamin, N, Guenneugues, M, Dauplais, M, Vita, C, Menez, A. | | Deposit date: | 1996-06-06 | | Release date: | 1997-01-11 | | Last modified: | 2020-01-15 | | Method: | SOLUTION NMR | | Cite: | NMR solution structure of a two-disulfide derivative of charybdotoxin: structural evidence for conservation of scorpion toxin alpha/beta motif and its hydrophobic side chain packing.
Biochemistry, 36, 1997
|
|
1BAI
 
 | | Crystal structure of Rous sarcoma virus protease in complex with inhibitor | | Descriptor: | N-[(2R)-2-({N~5~-[amino(iminio)methyl]-L-ornithyl-L-valyl}amino)-4-methylpentyl]-L-phenylalanyl-L-alpha-glutamyl-L-alanyl-L-norleucinamide, PROTEASE | | Authors: | Wu, J, Adomat, J.M, Ridky, T.W, Louis, J.M, Leis, J, Harrison, R.W, Weber, I.T. | | Deposit date: | 1998-04-17 | | Release date: | 1999-01-13 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural basis for specificity of retroviral proteases.
Biochemistry, 37, 1998
|
|
1BAJ
 
 | | HIV-1 CAPSID PROTEIN C-TERMINAL FRAGMENT PLUS GAG P2 DOMAIN | | Descriptor: | GAG POLYPROTEIN | | Authors: | Worthylake, D.K, Wang, H, Yoo, S, Sundquist, W.I, Hill, C.P. | | Deposit date: | 1998-04-17 | | Release date: | 1998-10-14 | | Last modified: | 2023-08-02 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structures of the HIV-1 capsid protein dimerization domain at 2.6 A resolution.
Acta Crystallogr.,Sect.D, 55, 1999
|
|
1BAK
 
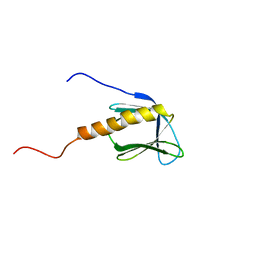 | |
1BAL
 
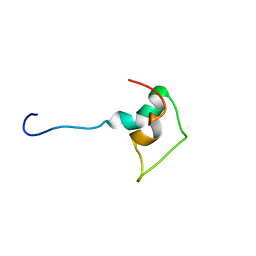 | |
1BAM
 
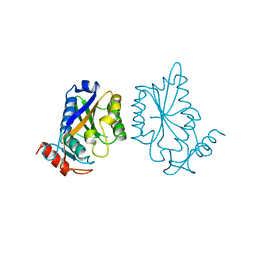 | |
1BAN
 
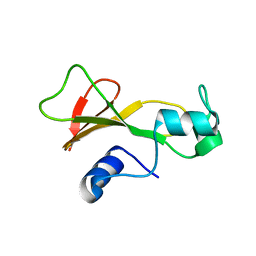 | |
1BAO
 
 | |
1BAP
 
 | |
1BAR
 
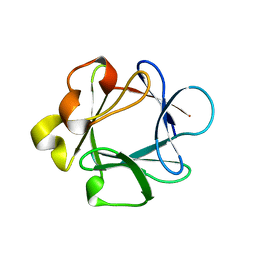 | | THREE-DIMENSIONAL STRUCTURES OF ACIDIC AND BASIC FIBROBLAST GROWTH FACTORS | | Descriptor: | ACIDIC FIBROBLAST GROWTH FACTOR | | Authors: | Zhu, X, Komiya, H, Chirino, A, Faham, S, Fox, G.M, Arakawa, T, Hsu, B.T, Rees, D.C. | | Deposit date: | 1992-09-29 | | Release date: | 1993-10-31 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Three-dimensional structures of acidic and basic fibroblast growth factors.
Science, 251, 1991
|
|
1BAS
 
 | |
1BAU
 
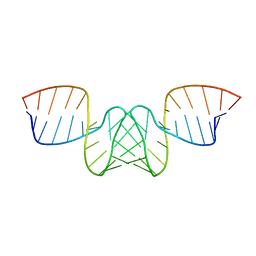 | | NMR STRUCTURE OF THE DIMER INITIATION COMPLEX OF HIV-1 GENOMIC RNA, MINIMIZED AVERAGE STRUCTURE | | Descriptor: | SL1 RNA DIMER | | Authors: | Mujeeb, A, Clever, J.L, Billeci, T.M, James, T.L, Parslow, T.G. | | Deposit date: | 1998-04-18 | | Release date: | 1999-04-27 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Structure of the dimer initiation complex of HIV-1 genomic RNA.
Nat.Struct.Biol., 5, 1998
|
|
1BAV
 
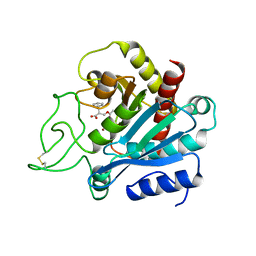 | |
1BAW
 
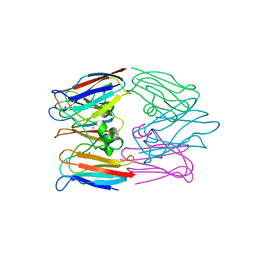 | | PLASTOCYANIN FROM PHORMIDIUM LAMINOSUM | | Descriptor: | COPPER (II) ION, PLASTOCYANIN, ZINC ION | | Authors: | Bond, C.S, Guss, J.M, Freeman, H.C, Wagner, M.J, Howe, C.J, Bendall, D.S. | | Deposit date: | 1998-04-19 | | Release date: | 1999-03-02 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | The structure of plastocyanin from the cyanobacterium Phormidium laminosum.
Acta Crystallogr.,Sect.D, 55, 1999
|
|
1BAX
 
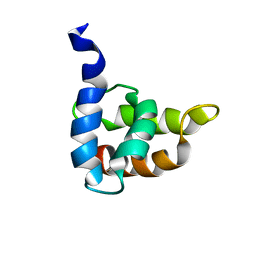 | | MASON-PFIZER MONKEY VIRUS MATRIX PROTEIN, NMR, AVERAGE STRUCTURE | | Descriptor: | M-PMV MATRIX PROTEIN | | Authors: | Conte, M.R, Klikova, M, Hunter, E, Ruml, T, Matthews, S. | | Deposit date: | 1998-04-20 | | Release date: | 1998-06-17 | | Last modified: | 2024-04-10 | | Method: | SOLUTION NMR | | Cite: | The three-dimensional solution structure of the matrix protein from the type D retrovirus, the Mason-Pfizer monkey virus, and implications for the morphology of retroviral assembly.
EMBO J., 16, 1997
|
|
1BAY
 
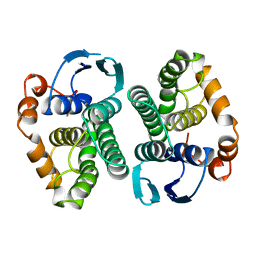 | | GLUTATHIONE S-TRANSFERASE YFYF CYS 47-CARBOXYMETHYLATED CLASS PI, FREE ENZYME | | Descriptor: | GLUTATHIONE S-TRANSFERASE CLASS PI | | Authors: | Vega, M.C, Coll, M. | | Deposit date: | 1996-11-02 | | Release date: | 1997-11-12 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The three-dimensional structure of Cys-47-modified mouse liver glutathione S-transferase P1-1. Carboxymethylation dramatically decreases the affinity for glutathione and is associated with a loss of electron density in the alphaB-310B region.
J.Biol.Chem., 273, 1998
|
|
1BAZ
 
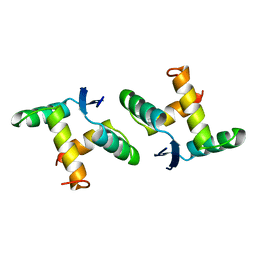 | |
1BB0
 
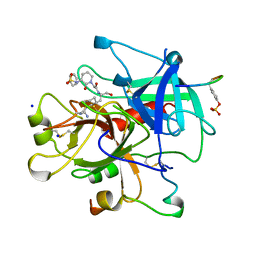 | | THROMBIN INHIBITORS WITH RIGID TRIPEPTIDYL ALDEHYDES | | Descriptor: | 2-{(3S)-3-[(benzylsulfonyl)amino]-2-oxopiperidin-1-yl}-N-{(2S)-1-[(3R)-1-carbamimidoylpiperidin-3-yl]-3-oxopropan-2-yl}acetamide, HIRUGEN, SODIUM ION, ... | | Authors: | Krishnan, R, Zhang, E, Hakansson, K, Arni, R.K, Tulinsky, A, Lim-Wilby, M.S.L, Levy, O.E, Semple, J.E, Brunck, T.K. | | Deposit date: | 1998-04-28 | | Release date: | 1999-04-27 | | Last modified: | 2023-08-02 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Highly selective mechanism-based thrombin inhibitors: structures of thrombin and trypsin inhibited with rigid peptidyl aldehydes.
Biochemistry, 37, 1998
|
|
1BB1
 
 | | CRYSTAL STRUCTURE OF A DESIGNED, THERMOSTABLE HETEROTRIMERIC COILED COIL | | Descriptor: | CHLORIDE ION, DESIGNED, THERMOSTABLE HETEROTRIMERIC COILED COIL | | Authors: | Nautiyal, S, Alber, T. | | Deposit date: | 1998-04-28 | | Release date: | 1999-02-02 | | Last modified: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of a designed, thermostable, heterotrimeric coiled coil.
Protein Sci., 8, 1999
|
|
1BB3
 
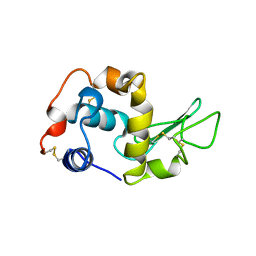 | |
1BB4
 
 | |
1BB5
 
 | |
