2F1N
 
 | |
2EUM
 
 | | Crystal structure of human Glycolipid Transfer Protein complexed with 8:0 Lactosylceramide | | Descriptor: | DECANE, Glycolipid transfer protein, N-OCTANE, ... | | Authors: | Malinina, L, Malakhova, M.L, Kanack, A.T, Abagyan, R, Brown, R.E, Patel, D.J. | | Deposit date: | 2005-10-28 | | Release date: | 2006-11-14 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | The liganding of glycolipid transfer protein is controlled by glycolipid acyl structure.
Plos Biol., 4, 2006
|
|
2GNA
 
 | | Crystal structure of UDP-GlcNAc inverting 4,6-dehydratase in complex with NADP and UDP-Gal | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, GALACTOSE-URIDINE-5'-DIPHOSPHATE, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, ... | | Authors: | Ishiyama, N, Creuzenet, C, Lam, J.S, Berghuis, A.M. | | Deposit date: | 2006-04-09 | | Release date: | 2006-05-09 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural Studies of FlaA1 from Helicobacter pylori Reveal the Mechanism for Inverting 4,6-Dehydratase Activity.
J.Biol.Chem., 281, 2006
|
|
2F22
 
 | |
2EVI
 
 | | Structure of a Ndt80-DNA complex (MSE mutant mA8T) | | Descriptor: | 5'-D(*AP*GP*TP*TP*AP*TP*TP*GP*TP*GP*TP*CP*GP*C)-3', 5'-D(*TP*GP*CP*GP*AP*CP*AP*CP*AP*AP*TP*AP*AP*C)-3', NDT80 protein | | Authors: | Lamoureux, J.S, Glover, J.N. | | Deposit date: | 2005-10-31 | | Release date: | 2006-03-21 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Principles of Protein-DNA Recognition Revealed in the Structural Analysis of Ndt80-MSE DNA Complexes.
Structure, 14, 2006
|
|
2GTM
 
 | | Mutated Mouse P38 MAP Kinase Domain in complex with Inhibitor PG-892579 | | Descriptor: | 8-(2-CHLOROPHENYLAMINO)-2-(2,6-DIFLUOROPHENYLAMINO)-9-ETHYL-9H-PURINE-1,7-DIIUM, Mitogen-activated protein kinase 14 | | Authors: | Walter, R.L, Mekel, M.J, Evdokimov, A.G, Pokross, M.E, Sabat, M. | | Deposit date: | 2006-04-28 | | Release date: | 2006-05-23 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The development of novel C-2, C-8, and N-9 trisubstituted purines as inhibitors of TNF-alpha production.
Bioorg.Med.Chem.Lett., 16, 2006
|
|
2GP7
 
 | | Estrogen Related Receptor-gamma ligand binding domain | | Descriptor: | CALCIUM ION, Estrogen-related receptor gamma | | Authors: | Wang, L, Zuercher, W.J, Consler, T.G, Lambert, M.H, Miller, A.B, Osband-miller, L.A, McKee, D.D, Willson, T.M, Nolte, R.T. | | Deposit date: | 2006-04-17 | | Release date: | 2006-09-26 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | X-ray crystal structures of the estrogen-related receptor-gamma ligand binding domain in three functional states reveal the molecular basis of small molecule regulation.
J.Biol.Chem., 281, 2006
|
|
2F54
 
 | | Directed evolution of human T cell receptor CDR2 residues by phage display dramatically enhances affinity for cognate peptide-MHC without increasing apparent cross-reactivity | | Descriptor: | Beta-2-microglobulin, Cancer/testis antigen 1B, HLA class I histocompatibility antigen, ... | | Authors: | Rizkallah, P.J, Jakobsen, B.K, Dunn, S.M, Sami, M. | | Deposit date: | 2005-11-25 | | Release date: | 2006-04-25 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Directed evolution of human T cell receptor CDR2 residues by phage display dramatically enhances affinity for cognate peptide-MHC without increasing apparent cross-reactivity.
Protein Sci., 15, 2006
|
|
2GQS
 
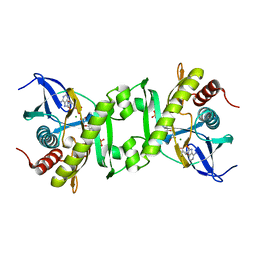 | | SAICAR Synthetase Complexed with CAIR-Mg2+ and ADP | | Descriptor: | 5-AMINO-1-(5-O-PHOSPHONO-BETA-D-RIBOFURANOSYL)-1H-IMIDAZOLE-4-CARBOXYLIC ACID, ADENOSINE-5'-DIPHOSPHATE, FORMIC ACID, ... | | Authors: | Ginder, N.D, Honzatko, R.B. | | Deposit date: | 2006-04-21 | | Release date: | 2006-05-16 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Nucleotide Complexes of Escherichia coli Phosphoribosylaminoimidazole Succinocarboxamide Synthetase.
J.Biol.Chem., 281, 2006
|
|
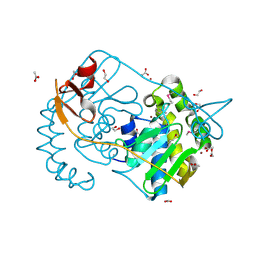
2GVI
 
 | |
2F7C
 
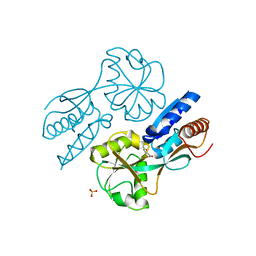 | | CatM effector binding domain with its effector cis,cis-muconate | | Descriptor: | (2Z,4Z)-HEXA-2,4-DIENEDIOIC ACID, HTH-type transcriptional regulator catM, SULFATE ION | | Authors: | Clark, T, Haddad, S, Ezezika, O, Neidle, E, Momany, C. | | Deposit date: | 2005-11-30 | | Release date: | 2006-10-31 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.162 Å) | | Cite: | Distinct Effector-binding Sites Enable Synergistic Transcriptional Activation by BenM, a LysR-type Regulator.
J.Mol.Biol., 367, 2007
|
|
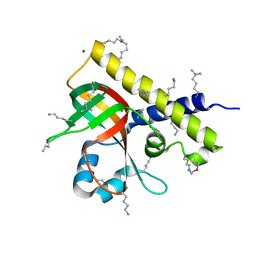
2F4I
 
 | |
2F8L
 
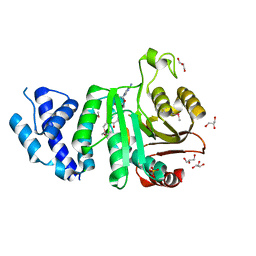 | |
2GZE
 
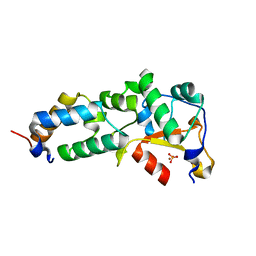 | | Crystal structure of the E9 DNase domain with a mutant immunity protein IM9 (Y55A) | | Descriptor: | Colicin-E9, Colicin-E9 immunity protein, PHOSPHATE ION, ... | | Authors: | Santi, P.S, Kolade, O.O, Kuhlmann, U.C, Kleanthous, C, Hemmings, A.M. | | Deposit date: | 2006-05-11 | | Release date: | 2008-03-18 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal Structure of the Complex of
the Colicin E9 DNase Domain with a Mutant Immunity Protein, IM9 (Y55A)
To be Published
|
|
2F6L
 
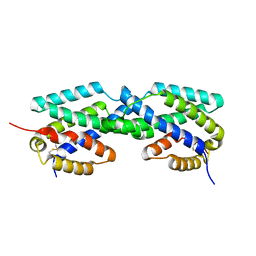 | | X-ray structure of Chorismate Mutase from Mycobacterium Tuberculosis | | Descriptor: | Chorismate mutase | | Authors: | Kim, S.K, Howard, A.J, Gilliland, G.L, Reddy, P.T, Ladner, J.E. | | Deposit date: | 2005-11-29 | | Release date: | 2005-12-13 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Biochemical and structural characterization of the secreted chorismate mutase (Rv1885c) from Mycobacterium tuberculosis H37Rv: an *AroQ enzyme not regulated by the aromatic amino acids.
J.Bacteriol., 188, 2006
|
|
2GZM
 
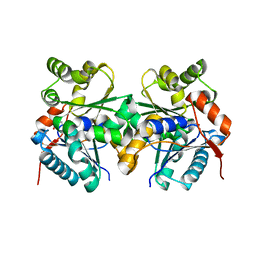 | | Crystal Structure of the Glutamate Racemase from Bacillus anthracis | | Descriptor: | D-GLUTAMIC ACID, Glutamate racemase | | Authors: | May, M, Santarsiero, B.D, Johnson, M.E, Mesecar, A.D. | | Deposit date: | 2006-05-11 | | Release date: | 2007-05-29 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | Structural and functional analysis of two glutamate racemase isozymes from Bacillus anthracis and implications for inhibitor design.
J.Mol.Biol., 371, 2007
|
|
2FAF
 
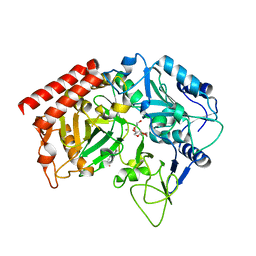 | | The structure of chicken mitochondrial PEPCK. | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, MANGANESE (II) ION, PENTAETHYLENE GLYCOL, ... | | Authors: | Holyoak, T, Sullivan, S.M, Nowak, T. | | Deposit date: | 2005-12-07 | | Release date: | 2006-06-27 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural Insights into the Mechanism of PEPCK Catalysis
Biochemistry, 45, 2006
|
|
2HDU
 
 | |
2FDK
 
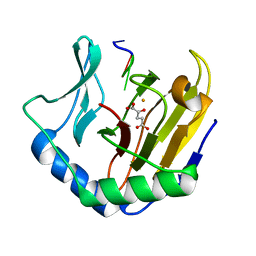 | | Crystal Structure of AlkB in complex with Fe(II), 2-oxoglutarate, and methylated trinucleotide T-meA-T (air 9 days) | | Descriptor: | 2-OXOGLUTARIC ACID, 5'-D(P*TP*(MA7)P*T)-3', Alkylated DNA repair protein alkB, ... | | Authors: | Yu, B, Benach, J, Edstrom, W.C, Gibney, B.R, Hunt, J.F, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2005-12-14 | | Release date: | 2006-02-21 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structures of catalytic complexes of the oxidative DNA/RNA repair enzyme AlkB.
Nature, 439, 2006
|
|
2FDC
 
 | |
2FFZ
 
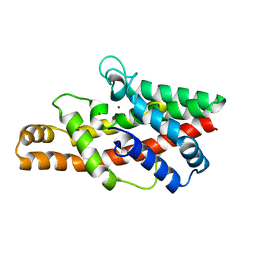 | |
2FLA
 
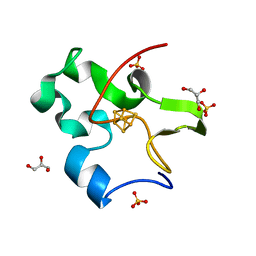 | | Structure of thereduced HiPIP from thermochromatium tepidum at 0.95 angstrom resolution | | Descriptor: | GLYCEROL, High-potential iron-sulfur protein, IRON/SULFUR CLUSTER, ... | | Authors: | Hunsicker-Wang, L.M, Stout, C.D, Fee, J.A. | | Deposit date: | 2006-01-05 | | Release date: | 2006-12-05 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (0.95 Å) | | Cite: | Geometric factors determine, in part, the electronic state of the 4Fe-4S cluster of HiPIP: a geometric, crystallographic, and theoretical study.
To be Published
|
|
2EUW
 
 | | Structure of a Ndt80-DNA complex (MSE mutant mA4T) | | Descriptor: | 5'-D(*AP*GP*TP*TP*TP*TP*TP*GP*AP*GP*TP*CP*GP*C)-3', 5'-D(*TP*GP*CP*GP*AP*CP*TP*CP*AP*AP*AP*AP*AP*C)-3', NDT80 protein | | Authors: | Lamoureux, J.S, Glover, J.N. | | Deposit date: | 2005-10-30 | | Release date: | 2006-03-21 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.68 Å) | | Cite: | Principles of Protein-DNA Recognition Revealed in the Structural Analysis of Ndt80-MSE DNA Complexes.
Structure, 14, 2006
|
|
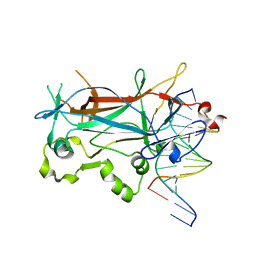
2ETR
 
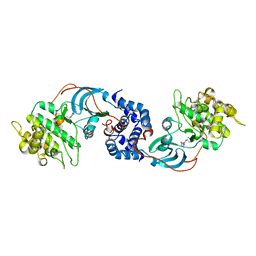 | | Crystal Structure of ROCK I bound to Y-27632 | | Descriptor: | (R)-TRANS-4-(1-AMINOETHYL)-N-(4-PYRIDYL) CYCLOHEXANECARBOXAMIDE, Rho-associated protein kinase 1 | | Authors: | Jacobs, M. | | Deposit date: | 2005-10-27 | | Release date: | 2005-11-08 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | The Structure of Dimeric ROCK I Reveals the Mechanism for Ligand Selectivity.
J.Biol.Chem., 281, 2006
|
|
2EZ8
 
 | | Pyruvate oxidase variant F479W in complex with reaction intermediate 2-lactyl-thiamin diphosphate | | Descriptor: | 3-[(4-AMINO-2-METHYLPYRIMIDIN-5-YL)METHYL]-2-(1-CARBOXY-1-HYDROXYETHYL)-5-(2-{[HYDROXY(PHOSPHONOOXY)PHOSPHORYL]OXY}ETHYL)-4-METHYL-1,3-THIAZOL-3-IUM, FLAVIN-ADENINE DINUCLEOTIDE, MAGNESIUM ION, ... | | Authors: | Wille, G, Meyer, D, Steinmetz, A, Hinze, E, Golbik, R, Tittmann, K. | | Deposit date: | 2005-11-10 | | Release date: | 2006-04-25 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.963 Å) | | Cite: | The catalytic cycle of a thiamin diphosphate enzyme examined by cryocrystallography.
Nat.Chem.Biol., 2, 2006
|
|
