9FOR
 
 | |
9FOF
 
 | |
9FMM
 
 | | Structure of human ACE2 in complex with a fluorinated small molecule inhibitor | | Descriptor: | (2~{S})-2-[[(2~{S})-3-[3-[(3-chloranyl-5-fluoranyl-phenyl)methyl]imidazol-4-yl]-1-oxidanyl-1-oxidanylidene-propan-2-yl]amino]-4-methyl-pentanoic acid, 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Benoit, R.B, Rodrigues, M.J, Wieser, M.M. | | Deposit date: | 2024-06-06 | | Release date: | 2024-07-17 | | Last modified: | 2024-08-14 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Development of radiofluorinated MLN-4760 derivatives for PET imaging of the SARS-CoV-2 entry receptor ACE2.
Eur J Nucl Med Mol Imaging, 2024
|
|
9FMD
 
 | |
9FLA
 
 | |
9FJK
 
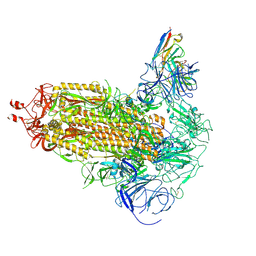 | | Omicron BA.1 Spike protein with neutralizing NTD specific mAb K501SP6 | | Descriptor: | K501SP6 Fv Heavy Chain, K501SP6 Fv Light Chain, Spike glycoprotein,Fibritin | | Authors: | Bjoernsson, K.H, Walker, M.R, Raghavan, S.S.R, Ward, A.B, Barfod, L.K. | | Deposit date: | 2024-05-31 | | Release date: | 2024-08-14 | | Method: | ELECTRON MICROSCOPY (2.84 Å) | | Cite: | Omicron BA.1 Spike protein with neutralizing NTD specific mAb K501SP6
To Be Published
|
|
9FJE
 
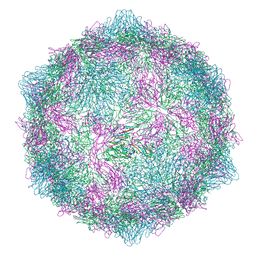 | | Expanded formalin inactivated CVB1 | | Descriptor: | Capsid protein VP1, Capsid protein VP2, Capsid protein VP3 | | Authors: | Plavec, Z, Butcher, S.J. | | Deposit date: | 2024-05-31 | | Release date: | 2024-07-31 | | Method: | ELECTRON MICROSCOPY (3.01 Å) | | Cite: | Comparison of structure and immunogenicity of CVB1-VLP and inactivated CVB1 vaccine candidates.
Res Sq, 2024
|
|
9FJD
 
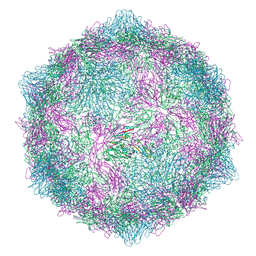 | | Expanded CVB1-VLP (Tween80) | | Descriptor: | Capsid protein VP1, Capsid protein VP2, Capsid protein VP3 | | Authors: | Plavec, Z, Butcher, S.J. | | Deposit date: | 2024-05-31 | | Release date: | 2024-07-31 | | Method: | ELECTRON MICROSCOPY (2.15 Å) | | Cite: | Comparison of structure and immunogenicity of CVB1-VLP and inactivated CVB1 vaccine candidates.
Res Sq, 2024
|
|
9FJC
 
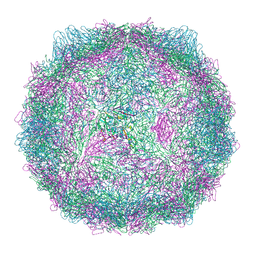 | | Compact CVB1-VLP (Tween80) | | Descriptor: | Capsid protein VP1, Capsid protein VP2, Capsid protein VP3, ... | | Authors: | Plavec, Z, Butcher, S.J. | | Deposit date: | 2024-05-30 | | Release date: | 2024-07-31 | | Method: | ELECTRON MICROSCOPY (2.15 Å) | | Cite: | Comparison of structure and immunogenicity of CVB1-VLP and inactivated CVB1 vaccine candidates.
Res Sq, 2024
|
|
9FIL
 
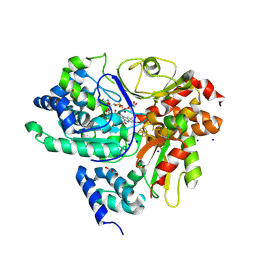 | | Crystal Structure of reduced NuoEF variant E222K(NuoF) from Aquifex aeolicus bound to NAD+ | | Descriptor: | 1-DEOXY-1-(7,8-DIMETHYL-2,4-DIOXO-3,4-DIHYDRO-2H-BENZO[G]PTERIDIN-1-ID-10(5H)-YL)-5-O-PHOSPHONATO-D-RIBITOL, CHLORIDE ION, FE2/S2 (INORGANIC) CLUSTER, ... | | Authors: | Wohlwend, D, Friedrich, T, Goeppert-Asadollahpour, S. | | Deposit date: | 2024-05-29 | | Release date: | 2024-07-17 | | Method: | X-RAY DIFFRACTION (2.54 Å) | | Cite: | Structural robustness of the NADH binding site in NADH:ubiquinone oxidoreductase (complex I).
Biochim Biophys Acta Bioenerg, 1865, 2024
|
|
9FIJ
 
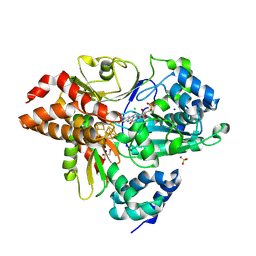 | | Crystal Structure of reduced NuoEF variant E222K(NuoF) from Aquifex aeolicus | | Descriptor: | 1-DEOXY-1-(7,8-DIMETHYL-2,4-DIOXO-3,4-DIHYDRO-2H-BENZO[G]PTERIDIN-1-ID-10(5H)-YL)-5-O-PHOSPHONATO-D-RIBITOL, CHLORIDE ION, FE2/S2 (INORGANIC) CLUSTER, ... | | Authors: | Wohlwend, D, Friedrich, T, Goeppert-Asadollahpour, S. | | Deposit date: | 2024-05-29 | | Release date: | 2024-07-17 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Structural robustness of the NADH binding site in NADH:ubiquinone oxidoreductase (complex I).
Biochim Biophys Acta Bioenerg, 1865, 2024
|
|
9FII
 
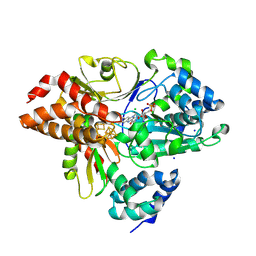 | | Crystal Structure of oxidized NuoEF variant E222K(NuoF) from Aquifex aeolicus | | Descriptor: | CHLORIDE ION, FE2/S2 (INORGANIC) CLUSTER, FLAVIN MONONUCLEOTIDE, ... | | Authors: | Wohlwend, D, Friedrich, T, Goeppert-Asadollahpour, S. | | Deposit date: | 2024-05-29 | | Release date: | 2024-07-17 | | Last modified: | 2024-08-14 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural robustness of the NADH binding site in NADH:ubiquinone oxidoreductase (complex I).
Biochim Biophys Acta Bioenerg, 1865, 2024
|
|
9FIH
 
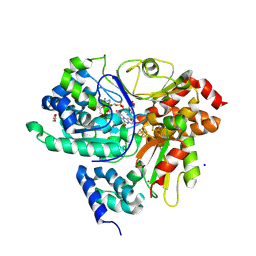 | | Crystal Structure of NuoEF variant P228R(NuoF) from Aquifex aeolicus bound to NADH under anoxic conditions after 10 min soaking | | Descriptor: | 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, 1-DEOXY-1-(7,8-DIMETHYL-2,4-DIOXO-3,4-DIHYDRO-2H-BENZO[G]PTERIDIN-1-ID-10(5H)-YL)-5-O-PHOSPHONATO-D-RIBITOL, FE2/S2 (INORGANIC) CLUSTER, ... | | Authors: | Wohlwend, D, Friedrich, T, Goeppert-Asadollahpour, S. | | Deposit date: | 2024-05-29 | | Release date: | 2024-07-17 | | Method: | X-RAY DIFFRACTION (2.08 Å) | | Cite: | Structural robustness of the NADH binding site in NADH:ubiquinone oxidoreductase (complex I).
Biochim Biophys Acta Bioenerg, 1865, 2024
|
|
9FIF
 
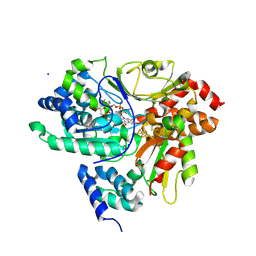 | | Crystal Structure of NuoEF variant P228R(NuoF) from Aquifex aeolicus bound to NADH under anoxic conditions | | Descriptor: | 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, 3[N-MORPHOLINO]PROPANE SULFONIC ACID, FE2/S2 (INORGANIC) CLUSTER, ... | | Authors: | Wohlwend, D, Friedrich, T, Goeppert-Asadollahpour, S. | | Deposit date: | 2024-05-29 | | Release date: | 2024-07-17 | | Method: | X-RAY DIFFRACTION (1.77 Å) | | Cite: | Structural robustness of the NADH binding site in NADH:ubiquinone oxidoreductase (complex I).
Biochim Biophys Acta Bioenerg, 1865, 2024
|
|
9FHP
 
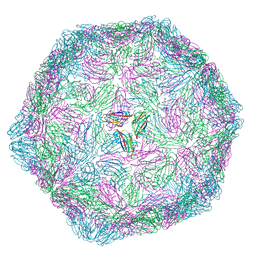 | |
9FH9
 
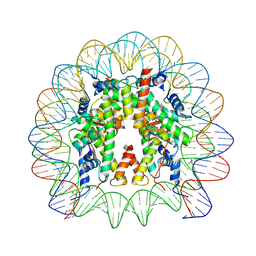 | |
9FGU
 
 | | SARS-CoV-2 (B.1.1.529/Omicron variant) Spike protein in complex with the single chain fragment scFv76-77 (focused refinement) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein,Fibritin, scFv76-77 single chain fragment | | Authors: | Berlinguer, M, Chaves-Sanjuan, A, Milazzo, F.M, Minenkova, O, De Santis, R, Bolognesi, M. | | Deposit date: | 2024-05-25 | | Release date: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (4.3 Å) | | Cite: | Structure of scFv76-77 in complex with SARS-CoV-2 Omicron Spike protein
To Be Published
|
|
9FGT
 
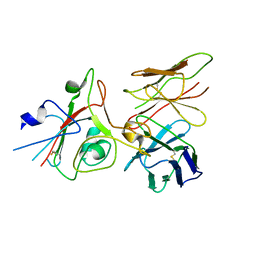 | | SARS-CoV-2 (B.1.1.529/Omicron variant) Spike protein in complex with the single chain fragment scFv76 (focused refinement) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Single chain fragment scFv76, Spike glycoprotein,Fibritin | | Authors: | Berlinguer, M, Chaves-Sanjuan, A, Milazzo, F.M, Minenkova, O, De Santis, R, Bolognesi, M. | | Deposit date: | 2024-05-25 | | Release date: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Structure of scFv76 in complex with SARS-CoV-2 Omicron Spike protein
To Be Published
|
|
9FGS
 
 | | SARS-CoV-2 (wuhan variant) Spike protein in complex with the single chain fragment scFv41N (focused refinement) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Single Chain fragment scFv41N, Spike glycoprotein,Fibritin | | Authors: | Berlinguer, M, Chaves-Sanjuan, A, Milazzo, F.M, Minenkova, O, De Santis, R, Bolognesi, M. | | Deposit date: | 2024-05-25 | | Release date: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | Structure of scFv41N in complex with SARS-CoV-2 Spike protein
To Be Published
|
|
9FGR
 
 | | SARS-CoV-2 (wuhan variant) Spike protein in complex with the single chain fragment scFv76-77 (focused refinement) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein, scFv76-77 single chain fragment | | Authors: | Berlinguer, M, Chaves-Sanjuan, A, Milazzo, F.M, Minenkova, O, De Santis, R, Bolognesi, M. | | Deposit date: | 2024-05-25 | | Release date: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | Structure of scFv76-77 in complex with SARS-CoV-2 Spike protein
To Be Published
|
|
9FGQ
 
 | |
9FGP
 
 | |
9FFH
 
 | | Native capsid of Rhodobacter microvirus Ebor computed with I4 symmetry | | Descriptor: | Major capsid protein | | Authors: | Bardy, P, MacDonald, C.I.W, Jenkins, H.T, Chechik, M, Hart, S.J, Turkenburg, J.P, Blaza, J.N, Fogg, P.C.M, Antson, A.A. | | Deposit date: | 2024-05-23 | | Release date: | 2024-06-12 | | Last modified: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | A stargate mechanism of Microviridae genome delivery unveiled by cryogenic electron tomography.
Biorxiv, 2024
|
|
9FFG
 
 | | Empty capsid of Rhodobacter microvirus Ebor computed with I4 symmetry | | Descriptor: | Major capsid protein | | Authors: | Bardy, P, MacDonald, C.I.W, Jenkins, H.T, Byrom, L, Chechik, M, Hart, S.J, Turkenburg, J.P, Blaza, J.N, Fogg, P.C.M, Antson, A.A. | | Deposit date: | 2024-05-23 | | Release date: | 2024-06-12 | | Last modified: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | A stargate mechanism of Microviridae genome delivery unveiled by cryogenic electron tomography.
Biorxiv, 2024
|
|
9FFF
 
 | | dsDNA-FANCD2-FANCI complex | | Descriptor: | DNA (32-MER), DNA (33-MER), Fanconi anemia complementation group I, ... | | Authors: | Alcon, P, Passmore, L.A. | | Deposit date: | 2024-05-23 | | Release date: | 2024-07-31 | | Method: | ELECTRON MICROSCOPY (3.68 Å) | | Cite: | dsDNA-FANCD2-FANCI complex
To Be Published
|
|
