1ALF
 
 | |
1BBA
 
 | |
1AGH
 
 | |
1SKH
 
 | |
1SMZ
 
 | |
2KYV
 
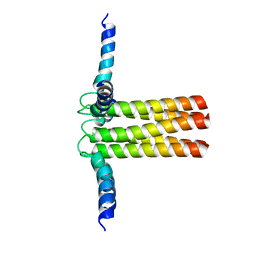 | | Hybrid solution and solid-state NMR structure ensemble of phospholamban pentamer | | Descriptor: | Phospholamban | | Authors: | Verardi, R, Shi, L, Traaseth, N.J, Veglia, G. | | Deposit date: | 2010-06-08 | | Release date: | 2011-05-11 | | Last modified: | 2024-05-01 | | Method: | SOLID-STATE NMR, SOLUTION NMR | | Cite: | Structural topology of phospholamban pentamer in lipid bilayers by a hybrid solution and solid-state NMR method.
Proc.Natl.Acad.Sci.USA, 108, 2011
|
|
1T3V
 
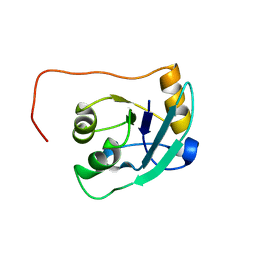 | | The NMR solution structure of TM1816 | | Descriptor: | conserved hypothetical protein | | Authors: | Columbus, L, Peti, W, Herrmann, T, Etazady, T, Klock, H, Lesley, S, Wuthrich, K, Joint Center for Structural Genomics (JCSG) | | Deposit date: | 2004-04-27 | | Release date: | 2004-12-14 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | NMR structure determination of the conserved hypothetical protein TM1816 from Thermotoga maritima.
Proteins, 60, 2005
|
|
1RHX
 
 | |
1T5Q
 
 | | Solution Structure of GIP(1-30)amide in TFE/Water | | Descriptor: | Gastric inhibitory polypeptide | | Authors: | Alana, I, Hewage, C.M, Malthouse, J.P.G, Parker, J.C, Gault, V.A, O'Harte, F.P.M. | | Deposit date: | 2004-05-05 | | Release date: | 2004-11-16 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | NMR structure of the glucose-dependent insulinotropic polypeptide fragment, GIP(1-30)amide.
Biochem.Biophys.Res.Commun., 325, 2004
|
|
1DJM
 
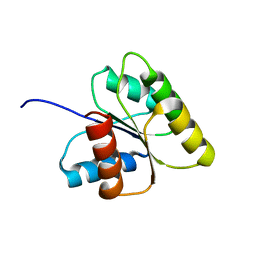 | | SOLUTION STRUCTURE OF BEF3-ACTIVATED CHEY FROM ESCHERICHIA COLI | | Descriptor: | CHEMOTAXIS PROTEIN Y | | Authors: | Cho, H.S, Lee, S.Y, Yan, D, Pan, X, Parkinson, J.S, Kustu, S, Wemmer, D.E, Pelton, J.G. | | Deposit date: | 1999-12-03 | | Release date: | 2000-04-05 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | NMR structure of activated CheY.
J.Mol.Biol., 297, 2000
|
|
1JO6
 
 | | Solution structure of the cytoplasmic N-terminus of the BK beta-subunit KCNMB2 | | Descriptor: | potassium large conductance calcium-activated channel, subfamily M, beta member 2 | | Authors: | Bentrop, D, Beyermann, M, Wissmann, R, Fakler, B. | | Deposit date: | 2001-07-27 | | Release date: | 2001-11-16 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | NMR structure of the "ball-and-chain" domain of KCNMB2, the beta 2-subunit of large conductance Ca2+- and voltage-activated potassium channels.
J.Biol.Chem., 276, 2001
|
|
1EIG
 
 | |
1EIH
 
 | |
1LD5
 
 | | STRUCTURE OF BPTI MUTANT A16V | | Descriptor: | PANCREATIC TRYPSIN INHIBITOR | | Authors: | Cierpicki, T, Otlewski, J. | | Deposit date: | 2002-04-08 | | Release date: | 2002-09-11 | | Last modified: | 2021-10-27 | | Method: | SOLUTION NMR | | Cite: | NMR structures of two variants of bovine pancreatic trypsin inhibitor (BPTI) reveal unexpected influence of mutations on protein structure and stability.
J.Mol.Biol., 321, 2002
|
|
1LD6
 
 | | STRUCTURE OF BPTI_8A MUTANT | | Descriptor: | PANCREATIC TRYPSIN INHIBITOR | | Authors: | Cierpicki, T, Otlewski, J. | | Deposit date: | 2002-04-08 | | Release date: | 2002-09-11 | | Last modified: | 2021-10-27 | | Method: | SOLUTION NMR | | Cite: | NMR structures of two variants of bovine pancreatic trypsin inhibitor (BPTI) reveal unexpected influence of mutations on protein structure and stability.
J.Mol.Biol., 321, 2002
|
|
1PFN
 
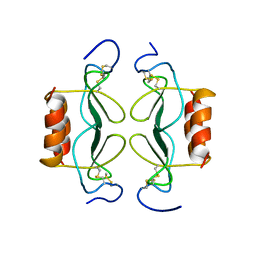 | | PF4-M2 CHIMERIC MUTANT WITH THE FIRST 10 N-TERMINAL RESIDUES OF R-PF4 REPLACED BY THE N-TERMINAL RESIDUES OF THE IL8 SEQUENCE. MODELS 16-27 OF A 27-MODEL SET. | | Descriptor: | PF4-M2 CHIMERA | | Authors: | Mayo, K.H, Roongta, V, Ilyina, E, Milius, R, Barker, S, Quinlan, C, La Rosa, G, Daly, T.J. | | Deposit date: | 1995-07-18 | | Release date: | 1996-01-29 | | Last modified: | 2021-11-03 | | Method: | SOLUTION NMR | | Cite: | NMR solution structure of the 32-kDa platelet factor 4 ELR-motif N-terminal chimera: a symmetric tetramer.
Biochemistry, 34, 1995
|
|
1PFM
 
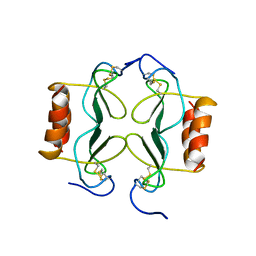 | | PF4-M2 CHIMERIC MUTANT WITH THE FIRST 10 N-TERMINAL RESIDUES OF R-PF4 REPLACED BY THE N-TERMINAL RESIDUES OF THE IL8 SEQUENCE. MODELS 1-15 OF A 27-MODEL SET. | | Descriptor: | PF4-M2 CHIMERA | | Authors: | Mayo, K.H, Roongta, V, Ilyina, E, Milius, R, Barker, S, Quinlan, C, La Rosa, G, Daly, T.J. | | Deposit date: | 1995-07-18 | | Release date: | 1996-01-29 | | Last modified: | 2021-11-03 | | Method: | SOLUTION NMR | | Cite: | NMR solution structure of the 32-kDa platelet factor 4 ELR-motif N-terminal chimera: a symmetric tetramer.
Biochemistry, 34, 1995
|
|
140D
 
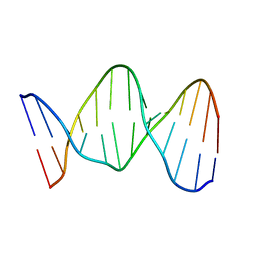 | | SOLUTION STRUCTURE OF A CONSERVED DNA SEQUENCE FROM THE HIV-1 GENOME: RESTRAINED MOLECULAR DYNAMICS SIMULATION WITH DISTANCE AND TORSION ANGLE RESTRAINTS DERIVED FROM TWO-DIMENSIONAL NMR SPECTRA | | Descriptor: | DNA (5'-D(*AP*GP*CP*TP*TP*GP*CP*CP*TP*TP*GP*AP*G)-3'), DNA (5'-D(*CP*TP*CP*AP*AP*GP*GP*CP*AP*AP*GP*CP*T)-3') | | Authors: | Mujeeb, A, Kerwin, S.M, Kenyon, G.L, James, T.L. | | Deposit date: | 1993-09-24 | | Release date: | 1994-04-30 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure of a conserved DNA sequence from the HIV-1 genome: restrained molecular dynamics simulation with distance and torsion angle restraints derived from two-dimensional NMR spectra.
Biochemistry, 32, 1993
|
|
142D
 
 | |
141D
 
 | |
108D
 
 | | THE SOLUTION STRUCTURE OF A DNA COMPLEX WITH THE FLUORESCENT BIS INTERCALATOR TOTO DETERMINED BY NMR SPECTROSCOPY | | Descriptor: | 1,1-(4,4,8,8-TETRAMETHYL-4,8-DIAZAUNDECAMETHYLENE)-BIS-4-3-METHYL-2,3-DIHYDRO-(BENZO-1,3-THIAZOLE)-2-METHYLIDENE)-QUINOLINIUM, DNA (5'-D(*CP*GP*CP*TP*AP*GP*CP*G)-3') | | Authors: | Spielmann, H.P, Wemmer, D.E, Jacobsen, J.P. | | Deposit date: | 1995-01-31 | | Release date: | 1995-06-03 | | Last modified: | 2024-03-13 | | Method: | SOLUTION NMR | | Cite: | Solution structure of a DNA complex with the fluorescent bis-intercalator TOTO determined by NMR spectroscopy.
Biochemistry, 34, 1995
|
|
1B5A
 
 | | RAT FERROCYTOCHROME B5 A CONFORMATION, NMR, 1 STRUCTURE | | Descriptor: | FERROCYTOCHROME B5, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Dangi, B, Sarma, S, Yan, C, Banville, D, Guiles, R.D. | | Deposit date: | 1998-04-06 | | Release date: | 1998-06-17 | | Last modified: | 2024-04-10 | | Method: | SOLUTION NMR | | Cite: | The origin of differences in the physical properties of the equilibrium forms of cytochrome b5 revealed through high-resolution NMR structures and backbone dynamic analyses.
Biochemistry, 37, 1998
|
|
1B5B
 
 | | RAT FERROCYTOCHROME B5 B CONFORMATION, NMR, 1 STRUCTURE | | Descriptor: | FERROCYTOCHROME B5, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Dangi, B, Sarma, S, Yan, C, Banville, D, Guiles, R.D. | | Deposit date: | 1998-04-06 | | Release date: | 1998-06-17 | | Last modified: | 2024-04-10 | | Method: | SOLUTION NMR | | Cite: | The origin of differences in the physical properties of the equilibrium forms of cytochrome b5 revealed through high-resolution NMR structures and backbone dynamic analyses.
Biochemistry, 37, 1998
|
|
1AC9
 
 | | SOLUTION STRUCTURE OF A DNA DECAMER CONTAINING THE ANTIVIRAL DRUG GANCICLOVIR: COMBINED USE OF NMR, RESTRAINED MOLECULAR DYNAMICS, AND FULL RELAXATION REFINEMENT, 6 STRUCTURES | | Descriptor: | DNA | | Authors: | Foti, M, Marshalko, S, Schurter, E, Kumar, S, Beardsley, G.P, Schweitzer, B.I. | | Deposit date: | 1997-02-17 | | Release date: | 1997-07-07 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure of a DNA decamer containing the antiviral drug ganciclovir: combined use of NMR, restrained molecular dynamics, and full relaxation matrix refinement.
Biochemistry, 36, 1997
|
|
1AFP
 
 | | SOLUTION STRUCTURE OF THE ANTIFUNGAL PROTEIN FROM ASPERGILLUS GIGANTEUS. EVIDENCE FOR DISULPHIDE CONFIGURATIONAL ISOMERISM | | Descriptor: | ANTIFUNGAL PROTEIN FROM ASPERGILLUS GIGANTEUS | | Authors: | Campos-Olivas, R, Bruix, M, Santoro, J, Lacadena, J, Del Pozo, A.M, Gavilanes, J.G, Rico, M. | | Deposit date: | 1994-11-11 | | Release date: | 1995-02-07 | | Last modified: | 2024-04-10 | | Method: | SOLUTION NMR | | Cite: | NMR solution structure of the antifungal protein from Aspergillus giganteus: evidence for cysteine pairing isomerism.
Biochemistry, 34, 1995
|
|
