2GL7
 
 | | Crystal Structure of a beta-catenin/BCL9/Tcf4 complex | | Descriptor: | B-cell lymphoma 9 protein, Beta-catenin, Transcription factor 7-like 2 | | Authors: | Sampietro, J. | | Deposit date: | 2006-04-04 | | Release date: | 2006-10-31 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal Structure of a beta-Catenin/BCL9/Tcf4 Complex.
Mol.Cell, 24, 2006
|
|
3CV0
 
 | | Structure of Peroxisomal Targeting Signal 1 (PTS1) binding domain of Trypanosoma brucei Peroxin 5 (TbPEX5)complexed to T. brucei Phosphoglucoisomerase (PGI) PTS1 peptide | | Descriptor: | 1,2-ETHANEDIOL, Peroxisome targeting signal 1 receptor PEX5, T. brucei PGI PTS1 peptide Ac-FNELSHL | | Authors: | Sampathkumar, P, Roach, C, Michels, P.A.M, Hol, W.G.J. | | Deposit date: | 2008-04-17 | | Release date: | 2008-06-24 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural Insights into the recognition of peroxisomal targeting signal 1 by Trypanosoma brucei peroxin 5.
J.Mol.Biol., 381, 2008
|
|
6WE6
 
 | | Camphor bound P450cam D251E structure | | Descriptor: | CAMPHOR, Camphor 5-monooxygenase, POTASSIUM ION, ... | | Authors: | Amaya, J.A, Poulos, T.L, Batabyal, D. | | Deposit date: | 2020-04-01 | | Release date: | 2020-07-08 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.16 Å) | | Cite: | Proton Relay Network in the Bacterial P450s: CYP101A1 and CYP101D1.
Biochemistry, 59, 2020
|
|
3DDK
 
 | | Coxsackievirus B3 3Dpol RNA Dependent RNA Polymerase | | Descriptor: | RNA polymerase B3 3Dpol, SODIUM ION, SULFATE ION | | Authors: | Campagnola, G, Weygandt, M.H, Scoggin, K.E, Peersen, O.B. | | Deposit date: | 2008-06-05 | | Release date: | 2008-09-23 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Crystal Structure of Coxsackievirus B3 3Dpol Highlights Functional Importance of Residue 5 in Picornaviral Polymerases
J.Virol., 82, 2008
|
|
3RBB
 
 | | HIV-1 NEF protein in complex with engineered HCK SH3 domain | | Descriptor: | 1,2-ETHANEDIOL, Protein Nef, Tyrosine-protein kinase HCK | | Authors: | Horenkamp, F.A, Schulte, A, Weyand, M, Geyer, M. | | Deposit date: | 2011-03-29 | | Release date: | 2011-04-27 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Conformation of the Dileucine-Based Sorting Motif in HIV-1 Nef Revealed by Intermolecular Domain Assembly.
Traffic, 12, 2011
|
|
1HY1
 
 | |
1G8E
 
 | | CRYSTAL STRUCTURE OF FLHD FROM ESCHERICHIA COLI | | Descriptor: | FLAGELLAR TRANSCRIPTIONAL ACTIVATOR FLHD | | Authors: | Campos, A, Zhang, R.G, Alkire, R.W, Matsumura, P, Westbrook, E.M. | | Deposit date: | 2000-11-17 | | Release date: | 2001-03-07 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of the global regulator FlhD from Escherichia coli at 1.8 A resolution.
Mol.Microbiol., 39, 2001
|
|
2Y0T
 
 | |
2Y0Q
 
 | |
2Y20
 
 | | The mechanisms of HAMP-mediated signaling in transmembrane receptors - the A291I mutant | | Descriptor: | UNCHARACTERIZED PROTEIN, ZINC ION | | Authors: | Zeth, K, Ferris, H.U, Hulko, M, Lupas, A.N. | | Deposit date: | 2010-12-12 | | Release date: | 2011-07-20 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | The Mechanisms of Hamp-Mediated Signaling in Transmembrane Receptors.
Structure, 19, 2011
|
|
2YGY
 
 | | Structure of wild type E. coli N-acetylneuraminic acid lyase in space group P21 crystal form II | | Descriptor: | CHLORIDE ION, N-ACETYLNEURAMINATE LYASE, PENTAETHYLENE GLYCOL | | Authors: | Campeotto, I, Nelson, A, Berry, A, Phillips, S.E.V, Pearson, A.R. | | Deposit date: | 2011-04-23 | | Release date: | 2012-04-04 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Pathological macromolecular crystallographic data affected by twinning, partial-disorder and exhibiting multiple lattices for testing of data processing and refinement tools.
Sci Rep, 8, 2018
|
|
3GLI
 
 | | Crystal Structure of the E. coli clamp loader bound to Primer-Template DNA and Psi Peptide | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, BERYLLIUM TRIFLUORIDE ION, DNA (5'-D(*CP*TP*GP*GP*CP*CP*TP*AP*TP*A)-3'), ... | | Authors: | Simonetta, K.R, Cantor, A.J, Kuriyan, J. | | Deposit date: | 2009-03-12 | | Release date: | 2009-05-26 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | The mechanism of ATP-dependent primer-template recognition by a clamp loader complex.
Cell(Cambridge,Mass.), 137, 2009
|
|
9C81
 
 | | X-ray crystal structure of AmpC beta-lactamase with inhibitor | | Descriptor: | (2R)-2-phenoxy-3-{[(1S,2S,4S)-spiro[bicyclo[2.2.1]heptane-7,1'-cyclopropane]-2-carbonyl]amino}propanoic acid, AmpC Beta-lactamase | | Authors: | Liu, F, Shoichet, B.K, Bassim, V. | | Deposit date: | 2024-06-11 | | Release date: | 2024-06-19 | | Last modified: | 2025-07-09 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | The impact of library size and scale of testing on virtual screening.
Nat.Chem.Biol., 21, 2025
|
|
9C84
 
 | |
9C6P
 
 | | X-ray crystal structure of AmpC beta-lactamase with inhibitor | | Descriptor: | 3-chloro-N-(5-chloro-2-methyl-1,3-benzothiazol-6-yl)-2-hydroxybenzene-1-sulfonamide, AmpC Beta-lactamase | | Authors: | Liu, F, Shoichet, B.K. | | Deposit date: | 2024-06-08 | | Release date: | 2024-06-19 | | Last modified: | 2025-07-09 | | Method: | X-RAY DIFFRACTION (1.663 Å) | | Cite: | The impact of library size and scale of testing on virtual screening.
Nat.Chem.Biol., 21, 2025
|
|
1XXH
 
 | | ATPgS Bound E. Coli Clamp Loader Complex | | Descriptor: | DNA polymerase III subunit gamma, DNA polymerase III, delta prime subunit, ... | | Authors: | Kazmirski, S.L, Podobnik, M, Weitze, T.F, O'Donnell, M, Kuriyan, J. | | Deposit date: | 2004-11-05 | | Release date: | 2004-12-07 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (3.45 Å) | | Cite: | Structural analysis of the inactive state of the Escherichia coli DNA polymerase clamp-loader complex
Proc.Natl.Acad.Sci.USA, 101, 2004
|
|
2Y7I
 
 | | Structural basis for high arginine specificity in Salmonella typhimurium periplasmic binding protein STM4351. | | Descriptor: | ACETATE ION, ARGININE, GLYCEROL, ... | | Authors: | Stamp, A.L, Owen, P, El Omari, K, Lockyer, M, Lamb, H.K, Charles, I.G, Hawkins, A.R, Stammers, D.K. | | Deposit date: | 2011-01-31 | | Release date: | 2011-05-11 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystallographic and Microcalorimetric Analyses Reveal the Structural Basis for High Arginine Specificity in the Salmonella Enterica Serovar Typhimurium Periplasmic Binding Protein Stm4351.
Proteins, 79, 2011
|
|
4Q9T
 
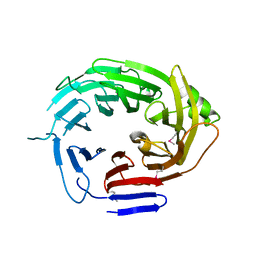 | |
4RJD
 
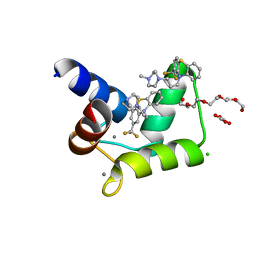 | | TFP bound in alternate orientations to calcium-saturated Calmodulin C-Domains | | Descriptor: | 10-[3-(4-METHYL-PIPERAZIN-1-YL)-PROPYL]-2-TRIFLUOROMETHYL-10H-PHENOTHIAZINE, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Feldkamp, M.D, Gakhar, L, Pandey, N, Shea, M.A. | | Deposit date: | 2014-10-08 | | Release date: | 2015-08-26 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Opposing orientations of the anti-psychotic drug trifluoperazine selected by alternate conformations of M144 in calmodulin.
Proteins, 83, 2015
|
|
4OU0
 
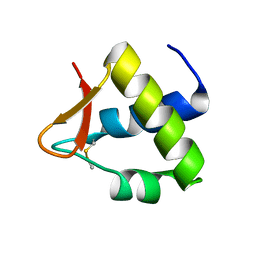 | |
4PAY
 
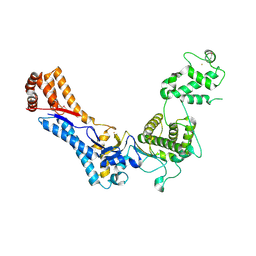 | | Crystal structure of an N-terminal fragment of the Legionella pneumophila effector protein SidC. | | Descriptor: | BARIUM ION, SidC, interaptin | | Authors: | Horenkamp, F.A, Mukherjee, S, Alix, E, Schauder, C.M, Hubber, A.M, Roy, C.R, Reinisch, K.M. | | Deposit date: | 2014-04-10 | | Release date: | 2014-06-25 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.77 Å) | | Cite: | Legionella pneumophila Subversion of Host Vesicular Transport by SidC Effector Proteins.
Traffic, 15, 2014
|
|
5W7H
 
 | | Supercharged arPTE variant R5 | | Descriptor: | Phosphotriesterase, ZINC ION | | Authors: | Campbell, E, Grant, J, Wang, Y, Sandhu, M, Williams, R.J, Nisbet, D.R, Perriman, A, Lupton, D, Jackson, C.J. | | Deposit date: | 2017-06-19 | | Release date: | 2019-01-23 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Hydrogel-Immobilized Supercharged Proteins
Adv Biosyst, 2018
|
|
8DB5
 
 | | Crystal structure of the GDP-D-glycero-4-keto-d-lyxo-heptose-3,5-epimerase from Campylobacter jejuni, serotype HS:15 | | Descriptor: | CHLORIDE ION, GDP-D-glycero-4-keto-d-lyxo-heptose-3,5-epimerase, GUANOSINE-5'-DIPHOSPHATE | | Authors: | Thoden, J.B, Ghosh, M.K, Xiang, D.F, Raushel, F.M, Holden, H.M. | | Deposit date: | 2022-06-14 | | Release date: | 2022-06-22 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | C3- and C3/C5-Epimerases Required for the Biosynthesis of the Capsular Polysaccharides from Campylobacter jejuni .
Biochemistry, 61, 2022
|
|
8DAK
 
 | | Crystal structure of the GDP-D-glycero-4-keto-d-lyxo-heptose-3-epimerase from Campylobacter jejuni, serotype HS:3 | | Descriptor: | CHLORIDE ION, GDP-D-glycero-4-keto-d-lyxo-heptose-3-epimerase, GUANOSINE-5'-DIPHOSPHATE, ... | | Authors: | Thoden, J.B, Ghosh, M.K, Xiang, D.F, Raushel, F.M, Holden, H.M. | | Deposit date: | 2022-06-13 | | Release date: | 2022-06-22 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | C3- and C3/C5-Epimerases Required for the Biosynthesis of the Capsular Polysaccharides from Campylobacter jejuni .
Biochemistry, 61, 2022
|
|
8DCO
 
 | | Crystal structure of the GDP-D-glycero-4-keto-D-lyxo-heptose-3,5-epimerase from Campylobacter jejuni, serotype HS:42 | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, GDP-D-glycero-4-keto-D-lyxo-heptose-3,5-epimerase, ... | | Authors: | Thoden, J.B, Xiang, D.F, Ghosh, M.K, Raushel, F.M, Holden, H.M. | | Deposit date: | 2022-06-17 | | Release date: | 2022-06-29 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | C3- and C3/C5-Epimerases Required for the Biosynthesis of the Capsular Polysaccharides from Campylobacter jejuni .
Biochemistry, 61, 2022
|
|
