1SWM
 
 | | X-RAY CRYSTAL STRUCTURE OF THE FERRIC SPERM WHALE MYOGLOBIN: IMIDAZOLE COMPLEX AT 2.0 ANGSTROMS RESOLUTION | | Descriptor: | AZIDE ION, MYOGLOBIN, PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | Rizzi, M, Ascenzi, P, Coda, A, Brunori, M, Bolognesi, M. | | Deposit date: | 1992-03-03 | | Release date: | 1994-01-31 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | X-ray crystal structure of the ferric sperm whale myoglobin: imidazole complex at 2.0 A resolution.
J.Mol.Biol., 217, 1991
|
|
1FKT
 
 | | SOLUTION STRUCTURE OF FKBP, A ROTAMASE ENZYME AND RECEPTOR FOR FK506 AND RAPAMYCIN | | Descriptor: | FK506 AND RAPAMYCIN-BINDING PROTEIN | | Authors: | Michnick, S.W, Rosen, M.K, Wandless, T.J, Karplus, M, Schreiber, S.L. | | Deposit date: | 1992-03-05 | | Release date: | 1994-01-31 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Solution structure of FKBP, a rotamase enzyme and receptor for FK506 and rapamycin.
Science, 252, 1991
|
|
1FKR
 
 | | SOLUTION STRUCTURE OF FKBP, A ROTAMASE ENZYME AND RECEPTOR FOR FK506 AND RAPAMYCIN | | Descriptor: | FK506 AND RAPAMYCIN-BINDING PROTEIN | | Authors: | Michnick, S.W, Rosen, M.K, Wandless, T.J, Karplus, M, Schreiber, S.L. | | Deposit date: | 1992-03-05 | | Release date: | 1994-01-31 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Solution structure of FKBP, a rotamase enzyme and receptor for FK506 and rapamycin.
Science, 252, 1991
|
|
1FKS
 
 | | SOLUTION STRUCTURE OF FKBP, A ROTAMASE ENZYME AND RECEPTOR FOR FK506 AND RAPAMYCIN | | Descriptor: | FK506 AND RAPAMYCIN-BINDING PROTEIN | | Authors: | Michnick, S.W, Rosen, M.K, Wandless, T.J, Karplus, M, Schreiber, S.L. | | Deposit date: | 1992-03-05 | | Release date: | 1994-01-31 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Solution structure of FKBP, a rotamase enzyme and receptor for FK506 and rapamycin.
Science, 252, 1991
|
|
1D66
 
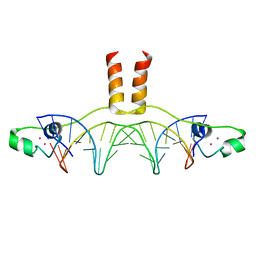 | | DNA RECOGNITION BY GAL4: STRUCTURE OF A PROTEIN/DNA COMPLEX | | Descriptor: | CADMIUM ION, DNA (5'-D(*CP*CP*GP*GP*AP*GP*GP*AP*CP*AP*GP*TP*CP*CP*TP*CP*C P*GP*G)-3'), DNA (5'-D(*CP*CP*GP*GP*AP*GP*GP*AP*CP*TP*GP*TP*CP*CP*TP*CP*C P*GP*G)-3'), ... | | Authors: | Marmorstein, R, Carey, M, Ptashne, M, Harrison, S.C. | | Deposit date: | 1992-03-06 | | Release date: | 1992-03-06 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | DNA recognition by GAL4: structure of a protein-DNA complex.
Nature, 356, 1992
|
|
2REB
 
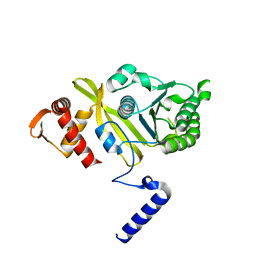 | |
1BBA
 
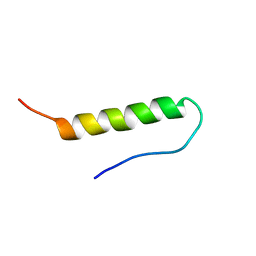 | |
1ENT
 
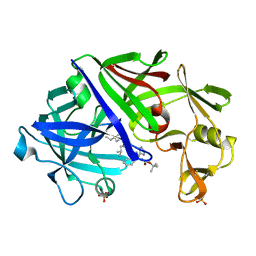 | |
1PLC
 
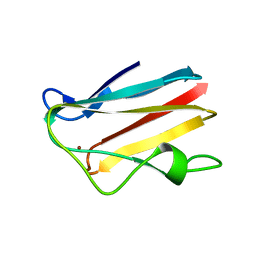 | |
4XIM
 
 | |
1SMR
 
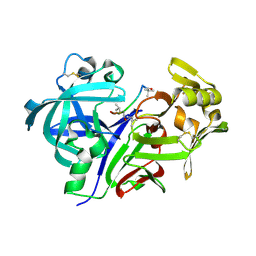 | |
1STP
 
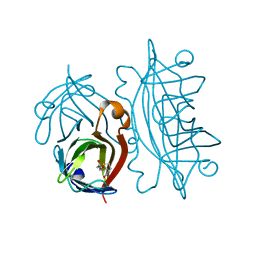 | |
1DR2
 
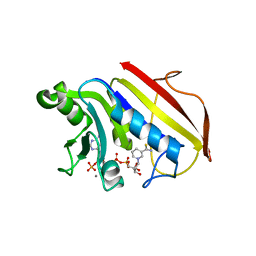 | | 2.3 ANGSTROMS CRYSTAL STRUCTURE OF CHICKEN LIVER DIHYDROFOLATE REDUCTASE COMPLEXED WITH THIONADP+ AND BIOPTERIN | | Descriptor: | 7-THIONICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, CALCIUM ION, DIHYDROFOLATE REDUCTASE | | Authors: | Mctigue, M.A, Davies /II, J.F, Kaufman, B.T, Xuong, N.-H, Kraut, J. | | Deposit date: | 1992-03-14 | | Release date: | 1993-10-31 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structures of chicken liver dihydrofolate reductase: binary thioNADP+ and ternary thioNADP+.biopterin complexes.
Biochemistry, 32, 1993
|
|
1DR4
 
 | | CRYSTAL STRUCTURES OF ORGANOMERCURIAL-ACTIVATED CHICKEN LIVER DIHYDROFOLATE REDUCTASE COMPLEXES | | Descriptor: | 7,8-DIHYDROBIOPTERIN, CALCIUM ION, DIHYDROFOLATE REDUCTASE, ... | | Authors: | Mctigue, M.A, Davies /II, J.F, Kaufman, B.T, Xuong, N.-H, Kraut, J. | | Deposit date: | 1992-03-14 | | Release date: | 1993-10-31 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal Structures of Organomercurial-Activated Chicken Liver Dihydrofolate Reductase Complexes
To be Published
|
|
1DR1
 
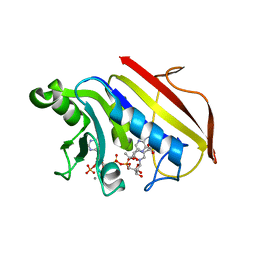 | | 2.2 ANGSTROMS CRYSTAL STRUCTURE OF CHICKEN LIVER DIHYDROFOLATE REDUCTASE COMPLEXED WITH NADP+ AND BIOPTERIN | | Descriptor: | 7,8-DIHYDROBIOPTERIN, CALCIUM ION, DIHYDROFOLATE REDUCTASE, ... | | Authors: | Mctigue, M.A, Davies /II, J.F, Kaufman, B.T, Xuong, N.-H, Kraut, J. | | Deposit date: | 1992-03-14 | | Release date: | 1993-10-31 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of chicken liver dihydrofolate reductase complexed with NADP+ and biopterin.
Biochemistry, 31, 1992
|
|
1DR6
 
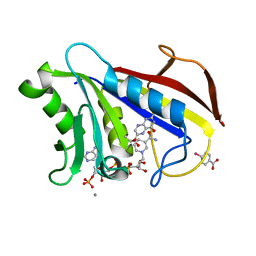 | | CRYSTAL STRUCTURES OF ORGANOMERCURIAL-ACTIVATED CHICKEN LIVER DIHYDROFOLATE REDUCTASE COMPLEXES | | Descriptor: | 7,8-DIHYDROBIOPTERIN, CALCIUM ION, DIHYDROFOLATE REDUCTASE, ... | | Authors: | Mctigue, M.A, Davies /II, J.F, Kaufman, B.T, Xuong, N.-H, Kraut, J. | | Deposit date: | 1992-03-14 | | Release date: | 1993-10-31 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal Structures of Organomercurial-Activated Chicken Liver Dihydrofolate Reductase Complexes
To be Published
|
|
1DR7
 
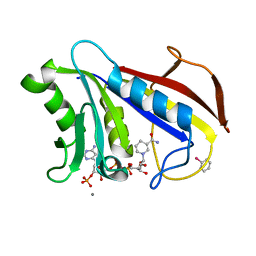 | | CRYSTAL STRUCTURES OF ORGANOMERCURIAL-ACTIVATED CHICKEN LIVER DIHYDROFOLATE REDUCTASE COMPLEXES | | Descriptor: | CALCIUM ION, DIHYDROFOLATE REDUCTASE, MERCURIBENZOIC ACID, ... | | Authors: | Mctigue, M.A, Davies /II, J.F, Kaufman, B.T, Xuong, N.-H, Kraut, J. | | Deposit date: | 1992-03-14 | | Release date: | 1993-10-31 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal Structures of Organomercurial-Activated Chicken Liver Dihydrofolate Reductase Complexes
To be Published
|
|
1DR5
 
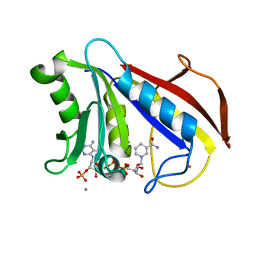 | | CRYSTAL STRUCTURES OF ORGANOMERCURIAL-ACTIVATED CHICKEN LIVER DIHYDROFOLATE REDUCTASE COMPLEXES | | Descriptor: | CALCIUM ION, DIHYDROFOLATE REDUCTASE, MERCURY (II) ION, ... | | Authors: | Mctigue, M.A, Davies /II, J.F, Kaufman, B.T, Xuong, N.-H, Kraut, J. | | Deposit date: | 1992-03-14 | | Release date: | 1993-10-31 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal Structures of Organomercurial-Activated Chicken Liver Dihydrofolate Reductase Complexes
To be Published
|
|
1DR3
 
 | | 2.3 ANGSTROMS CRYSTAL STRUCTURE OF CHICKEN LIVER DIHYDROFOLATE REDUCTASE COMPLEXED WITH THIONADP+ AND BIOPTERIN | | Descriptor: | 7,8-DIHYDROBIOPTERIN, 7-THIONICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, CALCIUM ION, ... | | Authors: | Mctigue, M.A, Davies /II, J.F, Kaufman, B.T, Xuong, N.-H, Kraut, J. | | Deposit date: | 1992-03-14 | | Release date: | 1993-10-31 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structures of chicken liver dihydrofolate reductase: binary thioNADP+ and ternary thioNADP+.biopterin complexes.
Biochemistry, 32, 1993
|
|
3LYT
 
 | |
4LYT
 
 | |
6LYT
 
 | |
5LYT
 
 | |
8FAB
 
 | |
1EPG
 
 | |
