1EDB
 
 | |
1EDP
 
 | |
1EGR
 
 | | SEQUENCE-SPECIFIC 1H N.M.R. ASSIGNMENTS AND DETERMINATION OF THE THREE-DIMENSIONAL STRUCTURE OF REDUCED ESCHERICHIA COLI GLUTAREDOXIN | | Descriptor: | GLUTAREDOXIN | | Authors: | Sodano, P, Xia, T.-H, Bushweller, J.H, Bjornberg, O, Holmgren, A, Billeter, M, Wuthrich, K. | | Deposit date: | 1991-10-08 | | Release date: | 1993-10-31 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Sequence-specific 1H n.m.r. assignments and determination of the three-dimensional structure of reduced Escherichia coli glutaredoxin.
J.Mol.Biol., 221, 1991
|
|
1BAN
 
 | |
1BCD
 
 | |
1AOZ
 
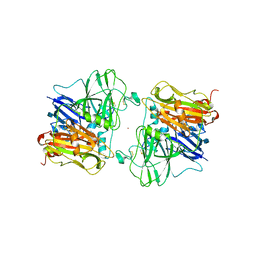 | | REFINED CRYSTAL STRUCTURE OF ASCORBATE OXIDASE AT 1.9 ANGSTROMS RESOLUTION | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, ASCORBATE OXIDASE, COPPER (II) ION, ... | | Authors: | Messerschmidt, A, Ladenstein, R, Huber, R. | | Deposit date: | 1992-01-08 | | Release date: | 1993-10-31 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Refined crystal structure of ascorbate oxidase at 1.9 A resolution.
J.Mol.Biol., 224, 1992
|
|
3CD4
 
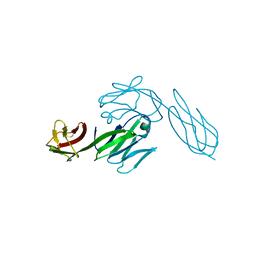 | |
3CI2
 
 | |
2LAL
 
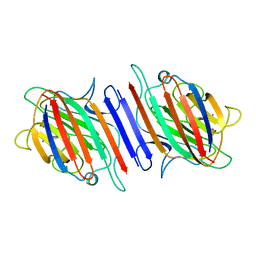 | | CRYSTAL STRUCTURE DETERMINATION AND REFINEMENT AT 2.3 ANGSTROMS RESOLUTION OF THE LENTIL LECTIN | | Descriptor: | CALCIUM ION, LENTIL LECTIN (ALPHA CHAIN), LENTIL LECTIN (BETA CHAIN), ... | | Authors: | Loris, R, Steyaert, J, Maes, D, Lisgarten, J, Pickersgill, R, Wyns, L. | | Deposit date: | 1993-06-10 | | Release date: | 1993-10-31 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural analysis of two crystal forms of lentil lectin at 1.8 A resolution.
Proteins, 20, 1994
|
|
1RTB
 
 | |
1RTA
 
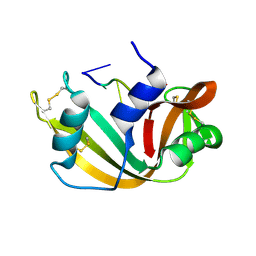 | |
1ERP
 
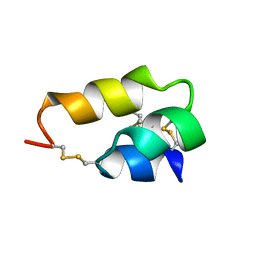 | | NUCLEAR MAGNETIC RESONANCE SOLUTION STRUCTURE OF THE PHEROMONE ER-10 FROM THE CILIATED PROTOZOAN EUPLOTES RAIKOVI | | Descriptor: | PHEROMONE ER-10 | | Authors: | Brown, L.R, Mronga, S, Bradshaw, R, Ortenzi, C, Luporini, P, Wuthrich, K. | | Deposit date: | 1992-12-02 | | Release date: | 1993-10-31 | | Last modified: | 2022-02-16 | | Method: | SOLUTION NMR | | Cite: | Nuclear magnetic resonance solution structure of the pheromone Er-10 from the ciliated protozoan Euplotes raikovi.
J.Mol.Biol., 231, 1993
|
|
1NIT
 
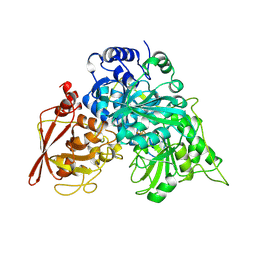 | | CRYSTAL STRUCTURE OF ACONITASE WITH TRANS-ACONITATE AND NITROCITRATE BOUND | | Descriptor: | ACONITASE, IRON/SULFUR CLUSTER, SULFATE ION | | Authors: | Lauble, H, Kennedy, M.C, Beinert, H, Stout, C.D. | | Deposit date: | 1993-01-17 | | Release date: | 1993-10-31 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Crystal structures of aconitase with trans-aconitate and nitrocitrate bound.
J.Mol.Biol., 237, 1994
|
|
1NIP
 
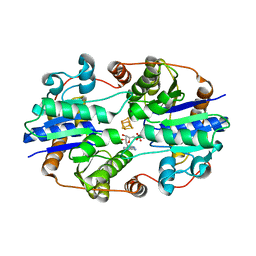 | | CRYSTALLOGRAPHIC STRUCTURE OF THE NITROGENASE IRON PROTEIN FROM AZOTOBACTER VINELANDII | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, IRON/SULFUR CLUSTER, MAGNESIUM ION, ... | | Authors: | Komiya, H, Georgiadis, M.M, Chakrabarti, P, Woo, D, Kornuc, J.J, Rees, D.C. | | Deposit date: | 1992-09-29 | | Release date: | 1993-10-31 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Crystallographic structure of the nitrogenase iron protein from Azotobacter vinelandii.
Science, 257, 1992
|
|
1RAQ
 
 | |
1RAZ
 
 | |
1RBE
 
 | |
1EAC
 
 | |
1EAB
 
 | |
1EAD
 
 | |
1INE
 
 | |
2TIR
 
 | | CRYSTAL STRUCTURE ANALYSIS OF A MUTANT ESCHERICHIA COLI THIOREDOXIN IN WHICH LYSINE 36 IS REPLACED BY GLUTAMIC ACID | | Descriptor: | COPPER (II) ION, THIOREDOXIN | | Authors: | Nikkola, M, Gleason, F.K, Fuchs, J.A, Eklund, H. | | Deposit date: | 1993-01-10 | | Release date: | 1993-10-31 | | Last modified: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure analysis of a mutant Escherichia coli thioredoxin in which lysine 36 is replaced by glutamic acid.
Biochemistry, 32, 1993
|
|
2HPD
 
 | | CRYSTAL STRUCTURE OF HEMOPROTEIN DOMAIN OF P450BM-3, A PROTOTYPE FOR MICROSOMAL P450'S | | Descriptor: | CYTOCHROME P450 BM-3, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Ravichandran, K.G, Boddupalli, S.S, Hasemann, C.A, Peterson, J.A, Deisenhofer, J. | | Deposit date: | 1993-09-16 | | Release date: | 1993-10-31 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of hemoprotein domain of P450BM-3, a prototype for microsomal P450's.
Science, 261, 1993
|
|
4RCR
 
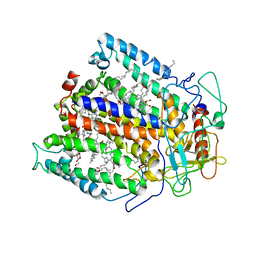 | | STRUCTURE OF THE REACTION CENTER FROM RHODOBACTER SPHAEROIDES R-26 AND 2.4.1: PROTEIN-COFACTOR (BACTERIOCHLOROPHYLL, BACTERIOPHEOPHYTIN, AND CAROTENOID) INTERACTIONS | | Descriptor: | BACTERIOCHLOROPHYLL A, BACTERIOPHEOPHYTIN A, FE (III) ION, ... | | Authors: | Komiya, H, Yeates, T.O, Chirino, A.J, Rees, D.C, Allen, J.P, Feher, G. | | Deposit date: | 1991-09-09 | | Release date: | 1993-10-31 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structure of the reaction center from Rhodobacter sphaeroides R-26 and 2.4.1: protein-cofactor (bacteriochlorophyll, bacteriopheophytin, and carotenoid) interactions.
Proc.Natl.Acad.Sci.USA, 85, 1988
|
|
1BAO
 
 | |
