1COK
 
 | |
1CRE
 
 | | CARDIOTOXIN II FROM TAIWAN COBRA VENOM, NAJA NAJA ATRA: STRUCTURE IN SOLUTION AND COMPARISION AMONG HOMOLOGOUS CARDIOTOXINS | | Descriptor: | CARDIOTOXIN II | | Authors: | Bhaskaran, R, Huang, C.C, Chang, K.D, Yu, C. | | Deposit date: | 1994-03-12 | | Release date: | 1994-05-31 | | Last modified: | 2017-11-29 | | Method: | SOLUTION NMR | | Cite: | Cardiotoxin II from Taiwan cobra venom, Naja naja atra. Structure in solution and comparison among homologous cardiotoxins.
J.Biol.Chem., 269, 1994
|
|
1CRF
 
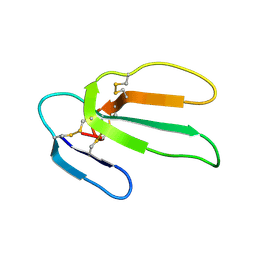 | | CARDIOTOXIN II FROM TAIWAN COBRA VENOM, NAJA NAJA ATRA: STRUCTURE IN SOLUTION AND COMPARISION AMONG HOMOLOGOUS CARDIOTOXINS | | Descriptor: | CARDIOTOXIN II | | Authors: | Bhaskaran, R, Huang, C.C, Chang, K.D, Yu, C. | | Deposit date: | 1994-03-12 | | Release date: | 1994-05-31 | | Last modified: | 2017-11-29 | | Method: | SOLUTION NMR | | Cite: | Cardiotoxin II from Taiwan cobra venom, Naja naja atra. Structure in solution and comparison among homologous cardiotoxins.
J.Biol.Chem., 269, 1994
|
|
1CIX
 
 | | THREE-DIMENSIONAL STRUCTURE OF ANTIMICROBIAL PEPTIDE TACHYSTATIN A ISOLATED FROM HORSESHOE CRAB | | Descriptor: | PROTEIN (TACHYSTATIN A) | | Authors: | Fujitani, N, Kawabata, S, Osaki, T, Kumaki, Y, Demura, M, Nitta, K, Kawano, K. | | Deposit date: | 1999-04-06 | | Release date: | 2002-05-01 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | Structure of the antimicrobial peptide tachystatin A.
J.Biol.Chem., 277, 2002
|
|
1ATE
 
 | |
1SSE
 
 | |
1D19
 
 | |
1VAZ
 
 | | Solution structures of the p47 SEP domain | | Descriptor: | NSFL1 cofactor p47 | | Authors: | Yuan, X, Simpson, P, Mckeown, C, Kondo, H, Uchiyama, K, Wallis, R, Dreveny, I, Keetch, C, Zhang, X, Robinson, C, Freemont, P, Matthews, S. | | Deposit date: | 2004-02-20 | | Release date: | 2004-04-06 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | Structure, dynamics and interactions of p47, a major adaptor of the AAA ATPase, p97.
Embo J., 23, 2004
|
|
1V89
 
 | | Solution Structure of the Pleckstrin Homology Domain of Human KIAA0053 Protein | | Descriptor: | Hypothetical protein KIAA0053 | | Authors: | Li, H, Tochio, N, Koshiba, S, Inoue, M, Hirota, H, Kigawa, T, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2003-12-29 | | Release date: | 2004-06-29 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | Solution Structure of the Pleckstrin Homology Domain of Human KIAA0053 Protein
To be Published
|
|
1D18
 
 | |
1RFO
 
 | | Trimeric Foldon of the T4 phagehead fibritin | | Descriptor: | whisker antigen control protein | | Authors: | Guthe, S, Kapinos, L, Moglich, A, Meier, S, Kiefhaber, T, Grzesiek, S. | | Deposit date: | 2003-11-10 | | Release date: | 2004-03-30 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Very fast folding and association of a trimerization domain from bacteriophage t4 fibritin.
J.Mol.Biol., 337, 2004
|
|
1V4Q
 
 | |
1V46
 
 | |
1V5T
 
 | | Solution Structure of the Ubiquitin-like Domain from Mouse Hypothetical 8430435I17Rik Protein | | Descriptor: | 8430435I17Rik Protein | | Authors: | Zhao, C, Kigawa, T, Tochio, N, Koshiba, S, Inoue, M, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2003-11-25 | | Release date: | 2004-05-25 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | Solution Structure of the Ubiquitin-like Domain from Mouse Hypothetical 8430435I17Rik Protein
To be Published
|
|
1BW3
 
 | |
1E0G
 
 | | LYSM Domain from E.coli MLTD | | Descriptor: | Membrane-bound lytic murein transglycosylase D | | Authors: | Bateman, A, Bycroft, M. | | Deposit date: | 2000-03-27 | | Release date: | 2000-06-21 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | The Structure of a Lysm Domain from E.Coli Membrane Bound Lytic Murein Transglycosylase D (Mltd)
J.Mol.Biol., 299, 2000
|
|
1AYI
 
 | | COLICIN E7 IMMUNITY PROTEIN IM7 | | Descriptor: | COLICIN E IMMUNITY PROTEIN 7 | | Authors: | Dennis, C.A, Pauptit, R.A, Wallis, R, James, R, Moore, G.R, Kleanthous, C. | | Deposit date: | 1997-11-04 | | Release date: | 1998-01-28 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | A structural comparison of the colicin immunity proteins Im7 and Im9 gives new insights into the molecular determinants of immunity-protein specificity.
Biochem.J., 333 ( Pt 1), 1998
|
|
1DSI
 
 | | Solution structure of a duocarmycin sa-indole-alkylated dna dupleX | | Descriptor: | 4-HYDROXY-6-(1H-INDOLE-2-CARBONYL)-8-METHYL-3,6,7,8-TETRAHYDRO-3,6-DIAZA-AS-INDACENE-2-CARBOXYLIC ACID METHYL ESTER, DNA (5'-D(*GP*AP*CP*TP*AP*AP*TP*TP*GP*AP*C)-3'), DNA (5'-D(*GP*TP*CP*AP*AP*TP*TP*AP*GP*TP*C)-3') | | Authors: | Schnell, J.R, Ketchem, R.R, Boger, D.L, Chazin, W.J. | | Deposit date: | 1998-07-29 | | Release date: | 1998-08-05 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Binding-Induced Activation of DNA Alkylation by Duocarmycin SA: Insights from the Structure of an Indole Derivative-DNA Adduct
J.Am.Chem.Soc., 121, 1999
|
|
1DVV
 
 | | SOLUTION STRUCTURE OF THE QUINTUPLE MUTANT OF CYTOCHROME C-551 FROM PSEUDOMONAS AERUGINOSA | | Descriptor: | CYTOCHROME C551, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Hasegawa, J, Uchiyama, S, Tanimoto, Y, Mizutani, M, Kobayashi, Y, Sambongi, Y, Igarashi, Y. | | Deposit date: | 2000-01-22 | | Release date: | 2000-11-29 | | Last modified: | 2024-04-10 | | Method: | SOLUTION NMR | | Cite: | Selected mutations in a mesophilic cytochrome c confer the stability of a thermophilic counterpart.
J.Biol.Chem., 275, 2000
|
|
1DEC
 
 | |
1DXN
 
 | |
1EV0
 
 | |
1P8A
 
 | |
2MY9
 
 | |
2MQI
 
 | |
