3PZY
 
 | |
3NF4
 
 | |
3PM6
 
 | |
3QK8
 
 | |
3Q8N
 
 | |
3P2Y
 
 | |
3P0X
 
 | |
3QKA
 
 | |
3QLJ
 
 | |
3QXI
 
 | |
3QDF
 
 | |
3QXZ
 
 | |
3QBP
 
 | |
6R7D
 
 | | Crystal structure of LTC4S in complex with AZ13690257 | | Descriptor: | (1~{S},2~{S})-2-[5-[cyclopropylmethyl(naphthalen-1-yl)amino]-4-methoxy-pyrimidin-2-yl]carbonylcyclopropane-1-carboxylic acid, DODECYL-BETA-D-MALTOSIDE, Leukotriene C4 synthase, ... | | Authors: | Kack, H, Ek, M. | | Deposit date: | 2019-03-28 | | Release date: | 2019-08-28 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Discovery of the Oral Leukotriene C4 Synthase Inhibitor (1S,2S)-2-({5-[(5-Chloro-2,4-difluorophenyl)(2-fluoro-2-methylpropyl)amino]-3-methoxypyrazin-2-yl}carbonyl)cyclopropanecarboxylic Acid (AZD9898) as a New Treatment for Asthma.
J.Med.Chem., 62, 2019
|
|
4GBR
 
 | |
1MRR
 
 | |
4GHK
 
 | |
4GK6
 
 | |
5WOU
 
 | |
1HS7
 
 | | VAM3P N-TERMINAL DOMAIN SOLUTION STRUCTURE | | Descriptor: | SYNTAXIN VAM3 | | Authors: | Dulubova, I, Yamaguchi, T, Wang, Y, Sudhof, T.C, Rizo, J. | | Deposit date: | 2000-12-24 | | Release date: | 2001-03-07 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Vam3p structure reveals conserved and divergent properties of syntaxins.
Nat.Struct.Biol., 8, 2001
|
|
2G2R
 
 | |
3VDI
 
 | | Structure of the FMO protein from Pelodictyon phaeum | | Descriptor: | BACTERIOCHLOROPHYLL A, TETRAETHYLENE GLYCOL, bacteriochlorophyll A protein | | Authors: | Tronrud, D.E, Larson, C.R, Seng, C.O, Lauman, L, Matthies, H.J, Wen, J, Blankenship, R.E, Allen, J.P. | | Deposit date: | 2012-01-05 | | Release date: | 2012-01-25 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | Reinterpretation of the electron density at the site of the eighth bacteriochlorophyll in the FMO protein from Pelodictyon phaeum.
Photosynth.Res., 112, 2012
|
|
7ZAK
 
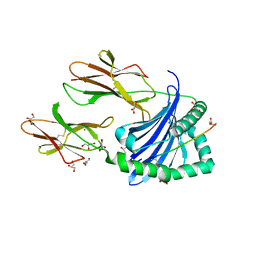 | | Crystal structure of HLA-DP (DPA1*02:01-DPB1*01:01) in complex with a peptide | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, GLYCEROL, MAGNESIUM ION, ... | | Authors: | Racle, J, Guillaume, P, Larabi, A, Lau, K, Pojer, F, Gfeller, D. | | Deposit date: | 2022-03-22 | | Release date: | 2023-03-29 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.62 Å) | | Cite: | Machine learning predictions of MHC-II specificities reveal alternative binding mode of class II epitopes.
Immunity, 56, 2023
|
|
1ESL
 
 | |
7ZFR
 
 | | Crystal structure of HLA-DP (DPA1*02:01-DPB1*01:01) in complex with a peptide bound in the reverse direction | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, MHC class II HLA-DP alpha chain (DPA1*02:01), MHC class II HLA-DP beta chain (DPB1*01:01), ... | | Authors: | Racle, J, Guillaume, P, Larabi, A, Lau, K, Pojer, F, Gfeller, D. | | Deposit date: | 2022-04-01 | | Release date: | 2023-04-12 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Machine learning predictions of MHC-II specificities reveal alternative binding mode of class II epitopes.
Immunity, 56, 2023
|
|
