1F4I
 
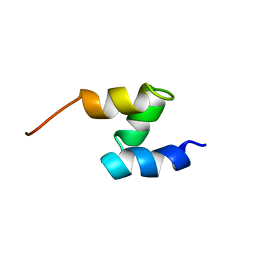 | | SOLUTION STRUCTURE OF THE HHR23A UBA(2) MUTANT P333E, DEFICIENT IN BINDING THE HIV-1 ACCESSORY PROTEIN VPR | | Descriptor: | UV EXCISION REPAIR PROTEIN PROTEIN RAD23 HOMOLOG A | | Authors: | Withers-Ward, E.S, Mueller, T.D, Chen, I.S, Feigon, J. | | Deposit date: | 2000-06-07 | | Release date: | 2000-12-20 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Biochemical and structural analysis of the interaction between the UBA(2) domain of the DNA repair protein HHR23A and HIV-1 Vpr.
Biochemistry, 39, 2000
|
|
1DV0
 
 | |
2UXV
 
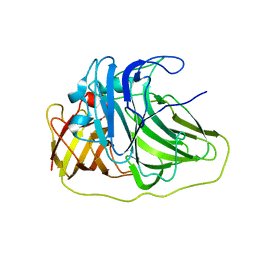 | | SufI Protein from Escherichia Coli | | Descriptor: | PROTEIN SUFI | | Authors: | Tarry, M.J, Roversi, P, Sargent, F, Berks, B.C, Lea, S.M. | | Deposit date: | 2007-03-30 | | Release date: | 2008-05-13 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.61 Å) | | Cite: | The Escherichia Coli Cell Division Protein and Model Tat Substrate Sufi (Ftsp) Localizes to the Septal Ring and Has a Multicopper Oxidase-Like Structure.
J.Mol.Biol., 386, 2009
|
|
2UXT
 
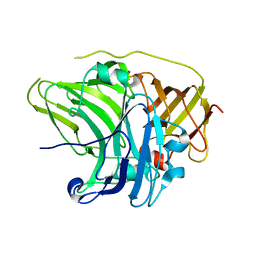 | | SufI Protein from Escherichia Coli | | Descriptor: | PROTEIN SUFI | | Authors: | Tarry, M.J, Roversi, P, Sargent, F, Berks, B.C, Lea, S.M. | | Deposit date: | 2007-03-29 | | Release date: | 2008-05-13 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The Escherichia Coli Cell Division Protein and Model Tat Substrate Sufi (Ftsp) Localizes to the Septal Ring and Has a Multicopper Oxidase-Like Structure.
J.Mol.Biol., 386, 2009
|
|
2VAE
 
 | | Fast maturing red fluorescent protein, DsRed.T4 | | Descriptor: | 1,2-ETHANEDIOL, RED FLUORESCENT PROTEIN | | Authors: | Strongin, D.E, Bevis, B, Khuong, N, Downing, M.E, Strack, R.L, Sundaram, K, Glick, B.S, Keenan, R.J. | | Deposit date: | 2007-08-31 | | Release date: | 2007-11-06 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.64 Å) | | Cite: | Structural Rearrangements Near the Chromophore Influence the Maturation Speed and Brightness of Dsred Variants.
Protein Eng.Des.Sel., 20, 2007
|
|
2VAD
 
 | | Monomeric red fluorescent protein, DsRed.M1 | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, RED FLUORESCENT PROTEIN, ... | | Authors: | Strongin, D.E, Bevis, B, Khuong, N, Downing, M.E, Strack, R.L, Sundaram, K, Glick, B.S, Keenan, R.J. | | Deposit date: | 2007-08-30 | | Release date: | 2007-11-06 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.59 Å) | | Cite: | Structural Rearrangements Near the Chromophore Influence the Maturation Speed and Brightness of Dsred Variants.
Protein Eng.Des.Sel., 20, 2007
|
|
2PSF
 
 | |
2PSD
 
 | |
2PSJ
 
 | |
2PSH
 
 | |
2PSE
 
 | |
5EB7
 
 | |
2JEE
 
 | | Xray structure of E. coli YiiU | | Descriptor: | CELL DIVISION PROTEIN ZAPB | | Authors: | Moller-Jensen, J, Gerdes, K, Lowe, J. | | Deposit date: | 2007-01-16 | | Release date: | 2008-02-05 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Novel Coiled-Coil Cell Division Factor Zapb Stimulates Z Ring Assembly and Cell Division.
Mol.Microbiol., 68, 2008
|
|
7QLL
 
 | |
7QLI
 
 | | Cis structure of rsKiiro at 290 K | | Descriptor: | GLYCEROL, SULFATE ION, rsKiiro | | Authors: | van Thor, J.J, Baxter, J.M. | | Deposit date: | 2021-12-20 | | Release date: | 2023-07-05 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.155 Å) | | Cite: | Optical control of ultrafast structural dynamics in a fluorescent protein.
Nat.Chem., 15, 2023
|
|
7QLK
 
 | |
9F8G
 
 | |
7QLJ
 
 | |
7Q6B
 
 | | mRubyFT/S148I, a mutant of blue-to-red fluorescent timer in its blue state | | Descriptor: | mRubyFT S148I, a mutant of blue-to-red fluorescent timer | | Authors: | Boyko, K.M, Nikolaeva, A.Y, Vlaskina, A.V, Dorovatovskii, P.V, Khrenova, M.G, Subach, O.M, Popov, V.O, Subach, F.M. | | Deposit date: | 2021-11-06 | | Release date: | 2023-04-12 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Combined Structural and Computational Study of the mRubyFT Fluorescent Timer Locked in Its Blue Form.
Int J Mol Sci, 24, 2023
|
|
7QLM
 
 | |
7QLN
 
 | | rsKiiro pump probe structure by TR-SFX | | Descriptor: | rsKiiro | | Authors: | van Thor, J.J. | | Deposit date: | 2021-12-20 | | Release date: | 2023-10-18 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Optical control of ultrafast structural dynamics in a fluorescent protein.
Nat.Chem., 15, 2023
|
|
5XG8
 
 | | Galectin-13/Placental Protein 13 variant R53H crystal structure | | Descriptor: | GLYCEROL, Galactoside-binding soluble lectin 13 | | Authors: | Wang, Y, Su, J.Y. | | Deposit date: | 2017-04-12 | | Release date: | 2018-01-31 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Galectin-13, a different prototype galectin, does not bind beta-galacto-sides and forms dimers via intermolecular disulfide bridges between Cys-136 and Cys-138
Sci Rep, 8, 2018
|
|
5Y03
 
 | | Galectin-13/Placental Protein 13 variant R53H crystal structure | | Descriptor: | Galactoside-binding soluble lectin 13 | | Authors: | Wang, Y, Su, J. | | Deposit date: | 2017-07-14 | | Release date: | 2018-01-31 | | Method: | X-RAY DIFFRACTION (2.12 Å) | | Cite: | Galectin-13, a different prototype galectin, does not bind beta-galacto-sides and forms dimers via intermolecular disulfide bridges between Cys-136 and Cys-138.
Sci Rep, 8, 2018
|
|
5WTS
 
 | |
2AR3
 
 | | E90A mutant structure of PlyL | | Descriptor: | PHOSPHATE ION, ZINC ION, prophage lambdaba02, ... | | Authors: | Low, L.Y, Yang, C, Perego, M, Osterman, A, Liddington, R.C. | | Deposit date: | 2005-08-19 | | Release date: | 2006-06-06 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure and lytic activity of a Bacillus anthracis prophage endolysin.
J.Biol.Chem., 280, 2005
|
|
